Description
Cyclic voltammetry (CV) is a type of electrochemical measurement which is applied with the Q-Module as quality control to (1) determine the oxidation and reduction peak potentials of Coenzyme Q in the specific experimental condition, (2) check the quality of the Q-Sensor, and (3) test the interference of chemicals used in the HRR assay with the Q-Sensor. In CV, the Q-Sensor with the three-electrode system is used to obtain information about the analyte (CoQ) by measuring the current (I) as the electric potential (V) between two of the electrodes is varied. In CV the electric potential between the glassy carbon (GC) and the Ag/AgCl reference electrode changes linearly versus time in cyclical phases, while the current is detected between GC and platinum electrode (Pt). The detected current is plotted versus the applied voltage to obtain the typical cyclic voltammogram trace (Figure 1). The presence of substances that are oxidized/reduced will result in current between GC and Pt, which can be seen as characteristic peaks in the voltammogram at a defined potential. The oxidation or the reduction peak potential values are used to set the GC (integrated into the Q-Sensor) for a separate experiment to measure the Q redox state of a biological sample. The oxidation and reduction peak potentials can be influenced by 1) the respiration medium, 2) the type of CoQ, 3) the polarization window, 4) the scan speed, 5) the number of cycles, 6) the concentration of the analyte (CoQ), and 7) the initial polarization voltage. <be>
Abbreviation: CV
Reference: Graham 2018 Standard operating procedures for cyclic voltammetry, Rich 1988 Glynn Res. Ph.
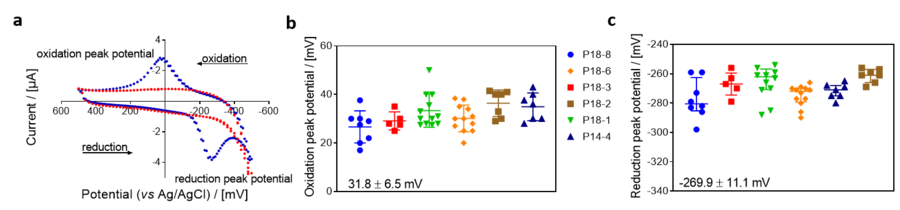
Figure 1. Cyclic voltammogram in the absence and presence of coenzyme Q2 (CoQ2) in non-stirred MiR05-Kit, at 37 °C by using the NextGen-O2k. Initial potential difference: +30 mV, polarization window: between -500 mV and +500 mV, scanning speed:100 mV/s, gain: 1. a: Blue plot: CV in the presence of CoQ2 (30 µM); red plot: background CV without CoQ2. Experiment: 2019-06-14_PQ2. Oxidation (b) and reduction (c) peak potentials of CoQ2 measured with cyclic voltammetry using different Q-Sensors. Codes of the Q-Sensors are next to the figure. Oxidation and reduction peak potentials are shown in the figure expressed as average ± SD [mV]; n=51.
The following parameters are taken into account when measuring CV:
• Initial polarization voltage is the potential where the scanning starts. In order to avoid coating of the GCE, it must be close to the peak potential (Graham 2018). In the case of CoQ2, +30 mV was used as an initial potential as it is close to the oxidation peak potential.
• Polarization window: The narrowest possible range of potentials should be applied during the CV scanning. High and low potentials might lead to chemical modification or coating of the GCE, therefore, wider than necessary polarization windows should be avoided (Graham 2018). Any type of modification of GCE can inhibit the electron transfer on the surface of it. In the case of CoQ2, -500 mV and +500 mV were chosen as a potential window.
• Number of CV cycles: Although after one cycle (potential changes from -500 mV to +500 mV and then back to -500 mV) well-defined peaks for oxidation and reduction potentials are observed, it is recommended to run more cycles to check whether additional peaks are detected or the shape of the oxidation and reduction peak potential changes over the cycles owing to side-reactions. In the Q-Module case, a total of five cycles are performed.
• Scan speed: The scanning speed should support free diffusion of CoQ2. If the scanning speed is very slow, CoQ2 might be transported to and from the electrode surface via other processes than diffusion (Graham 2018). If the scanning speed is too fast, it leads to double layer charging current, which comes from the rearrangement of solution molecules at the surface of GCE as a result of the changing electrode potential and results in high baseline current that obscure features in a CV (Graham 2018). To avoid any of these side reactions and provide free diffusion for CoQ2, 100 mV/s was applied as a scanning speed.
• Non-stirred solution: Stirring of the solution should be avoided during CV. Upon stirring if there is only quinone (oxidized CoQ), only a wave of quinone reduction is visible and the wave of quinol (reduced CoQ) oxidation cannot be observed, because the quinol is stirred off from the surface of GCE (Peter R Rich, personal communication).
• Optimal concentration of CoQ2 for CV: For CV, the lowest possible concentration of CoQ2 should be applied which gives well-defined oxidation and reduction peak potentials. In MiR05-Kit, 30 µM CoQ2 was ideal for CV, because lower than 30 µM of CoQ2 did not give detectable peaks at gain of 1 V/µA, whereas using higher than ~ 90 µM of CoQ2 the limit of detection was reached.
• Temperature slightly influences the peak potential values; therefore, CV is performed at experimental temperature.
Cyclic voltammetry is part of the Q-Module and the NextGen-O2k project
- The Q-Module allows for monitoring of the redox state of electron transfer-reactive coenzyme Q at the Q-junction using the specific Q-Stoppers with the integrated three-electrode system and the modified electronics of the NextGen-O2k. Cyclic voltammetry is used for quality control and for defining the polarization voltage applied during Q-redox measurements.
- Reference:
- Komlódi T, Cardoso LHD, Doerrier C, Moore AL, Rich PR, Gnaiger E (2021) Coupling and pathway control of coenzyme Q redox state and respiration in isolated mitochondria. Bioenerg Commun 2021.3. https://doi.org/10.26124/bec:2021-0003
- Reference:
Communicated by Komlodi T, Cardoso LHD (last update 2021-02-09)
- Bioblast links: Q - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coenzyme Q
- » Coenzyme Q
- » Quinone, Ubiquinone Q; oxidized
- » Quinol, Ubiquinol QH2; reduced
- » Semiquinone
- » Coenzyme Q2
- » Q-redox state
- » Q-pools
- Coenzyme Q
- Mitochondrial pathways, respiratory Complexes, and Q
- » Q-cycle
- » Q-junction
- » Convergent electron flow
- » NS-pathway
- » FNS
- » FNSGp
- Mitochondrial pathways, respiratory Complexes, and Q
- NextGen-O2k and Q-Module