Iannetti 2015 Abstract MiP2015
| High-Content Screening of mitochondrial morphofunction in living cells. |
Link:
Iannetti E, Smeitink J, Beyrath J, Willems P, Koopman WJ (2015)
Event: MiP2015
Mitochondrial morphology and functionality (“morphofunction”) have often been described in a simplistic binary manner as “normal” or “aberrant” in various pathophysiological conditions. However mitochondrial morphofucntional phenotypes, depending on a balance of countless factors, are not so easy to label. Mitochondrial physio-pathology usually presents a continuum of morphofunctional states [1].
To properly describe this state continuum, simultaneous monitoring of multiple parameters is required. Therefore, here we present an integrated strategy allowing quantification of mitochondrial morphofunction in single intact living cells using multivariate data sets rather than single measurements. To this end, three fluorescent reporter molecules are combined into a multispectral fluorescence microscopy assay. In contrast to manual classification the presented approach allows combined high-content and high-throughput analysis of multi-well plates (High-content Screening). The automated analysis of large data sets drastically reduces bias, provides strong statistical power, and enables reliable analysis between various cell lines and conditions.
The proposed HCS technology is able to discriminate drug-induced modulation of mitochondrial phenotypes. Moreover, the assay can clearly individuate and quantify pathophysiological morphofunctional phenotypes of cell lines carrying or not carrying pathogenic mutations.
Therefore, the technology is proposed as an ideal platform to perform library drug screening [2] and mode-of action studies but also to address fundamental questions in mitochondrial research.
• O2k-Network Lab: NL Nijmegen Koopman WJ
Labels: MiParea: Instruments;methods, mt-Medicine, Pharmacology;toxicology
Stress:Mitochondrial disease
Event: B1, Oral
MiP2015
Affiliations
1-Dept Biochem, Radboud Inst Mol Life Sc, Radboud Univ Med Center; 2-Khondrion BV; 3-Dept Pediatrics, Nijmegen Centre Mitoch Disorders, Radboud Univ Med Center; 4-Centre Systems Biol Bioenergetics, Radboud Univ Med Center; 4-Khondrion BV, Nijmegen, The Netherlands. - [email protected]
Figures
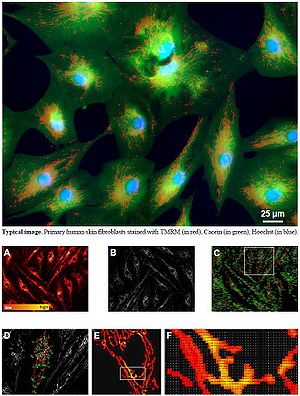
Image processing and data extraction. (A) TMRM RAW image. Fluorescence intensity is color-coded using a red to yellow scale. (B) MASKED image obtained by applying an automated image processing algorithm. (C) Computer-assisted identification of mitochondrial objects. Each object is described by 31 morphological and functional escriptors; these parameters are automatically extracted and provide a multidimensional phenotypic quantification of the mitochondrial network morphology and functionality. (D) Magnification of a region of interest in panel C. (E) Magnification of a region of interest in panel D. (F) Bitmap of a region of interest in panel E.
References and acknowledgements
- Iannetti EF, Willems PH, Pellegrini M, Beyrath J, Smeitink JA, Blanchet L, Koopman WJ (2015) Toward high-content screening of mitochondrial morphology and membrane potential in living cells. Int J Biochem Cell Biol 63:66-70.
- Blanchet L, Smeitink JA, van Emst-de Vries SE, Vogels C, Pellegrini M, Jonckheere AI, Rodenburg RJ, Buydens LM, Beyrath J, Willems PH, Koopman WJ (2015) Quantifying small molecule phenotypic effects using mitochondrial morpho-functional fingerprinting and machine learning. Sci Rep 5:8035.