Sazanov 2015 Nat Rev Mol Cell Biol
| Sazanov LA (2015) A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol 16:375-88. https://doi.org/10.1038/nrm3997 |
Sazanov Leonid A (2015) Nat Rev Mol Cell Biol
Abstract: The mitochondrial respiratory chain, also known as the electron transport chain (ETC), is crucial to life, and energy production in the form of ATP is the main mitochondrial function. Three proton-translocating enzymes of the ETC, namely complexes I, III and IV, generate proton motive force, which in turn drives ATP synthase (complex V). The atomic structures and basic mechanisms of most respiratory complexes have previously been established, with the exception of complex I, the largest complex in the ETC. Recently, the crystal structure of the entire complex I was solved using a bacterial enzyme. The structure provided novel insights into the core architecture of the complex, the electron transfer and proton translocation pathways, as well as the mechanism that couples these two processes.
• Bioblast editor: Gnaiger E
Hydrogen ion ambiguities in the electron transfer system
Communicated by Gnaiger E (2023-10-08) last update 2023-11-10
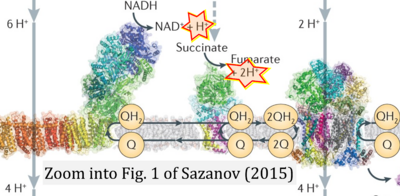
- Electron (e-) transfer linked to hydrogen ion (hydron; H+) transfer is a fundamental concept in the field of bioenergetics, critical for understanding redox-coupled energy transformations.
- However, the current literature contains inconsistencies regarding H+ formation on the negative side of bioenergetic membranes, such as the matrix side of the mitochondrial inner membrane, when NADH is oxidized during oxidative phosphorylation (OXPHOS). Ambiguities arise when examining the oxidation of NADH by respiratory Complex I or succinate by Complex II.
- Oxidation of NADH or succinate involves a two-electron transfer of 2{H++e-} to FMN or FAD, respectively. Figures indicating a single electron e- transferred from NADH or succinate lack accuracy.
- The oxidized NAD+ is distinguished from NAD indicating nicotinamide adenine dinucleotide independent of oxidation state.
- NADH + H+ → NAD+ +2{H++e-} is the oxidation half-reaction in this H+-linked electron transfer represented as 2{H++e-} (Gnaiger 2023). Putative H+ formation shown as NADH → NAD+ + H+ conflicts with chemiosmotic coupling stoichiometries between H+ translocation across the coupling membrane and electron transfer to oxygen. Ensuring clarity in this complex field is imperative to tackle the apparent ambiguity crisis and prevent confusion, particularly in light of the increasing number of interdisciplinary publications on bioenergetics concerning diagnostic and clinical applications of OXPHOS analysis.
Cited by
- Gnaiger E et al ― MitoEAGLE Task Group (2020) Mitochondrial physiology. Bioenerg Commun 2020.1. doi:10.26124/bec:2020-0001.v1.
Labels:
Enzyme: Complex I Regulation: ATP production, Coupling efficiency;uncoupling
BEC 2020.1