Talk:The protonmotive force and respiratory control
From Bioblast
Revision as of 17:56, 29 August 2017 by Gnaiger Erich (talk | contribs)
Work flow
- 2017-08-29 Marco Spinazzi:
- As discussed in a previous workshop together, I believe this is a great initiative since so much confusion is in the general science community, especially since the recent resurrection of the interest for mitochondria from different specialists not previously involved in mitochondrial research. My very personal point of view is that the manuscript should be a bit restyled in order to focus on LESS but VERY clear concepts that can be easily digested by non specialists, in order to have a larger audience and impact, otherwise the paper will mainly talk to specialist, many of which are at least partly familiar with these informations. Please have a look how effectively these papers addressed methodological issues on cell death and autophagy. They are very easy to read and had huge impact. I would personally try to simplify the key concepts, cut energically overclassifications and formulae, limit historical citations, and possibly add a couple of experiments to illustrate the different fluxes. Limit abbreviations to a minimum. And more clearly explain what are OXPHOS and ETS, what it relies on, and what it means if 1 or both are decreased. Possibly with an illustrative trace. For the rest, with this first read, I have some other, more circumscribed comments:
- -Page 6: I don’t understand where the content of page 6 goes. The IM separates the IMS from matrix. In some place of the manuscript I would more clearly states what the Respiratory chain includes (CI-CIV) compared to OXPHOS system (Respiratory chain + CV). Also not enough highlighted that OXPHOS rely also on Krebs. - (EG: see Fig. 1).
- In page 6, other important mitochondrial components in addition to OXPHOS are mentioned, but many others have not: calcium metabolism, lipid, Coq10 and steroid biosynthesis, vitamin metabolism, etc…. Maybe just simpler to mention that the mitochondrial proteome comprises more than 1200 proteins (Mitocharta), mostly encoded by nDNA, with a fantastic variety of functions, many of which still under investigation… (EG: added).
- -I personally do not like ETS, and prefer ETC=electron transfer capacity.
- EG: ETS - system: this is the structural and functional unit; Capacity: this relates to flux. A distinction is required.
- -page 8: I don’t like to define mitochondrial preparations tissue homogenates, permeabilized fibers/tissues, but rather would define as they are : tissue homogenates, permeabilized fibers/tissues. Mitochondrial preparations are in my opinion mitochondrial enriched fraction or purified mitochondria (either by Percoll, sucrose or analogues). I don’t agree on the fact that these preparation largely maintain the mitochondrial structure, and very often not even the function. On the contrary most of these traetments introduce modifications in both, that can often be even quite drastic. This is one of the main limitation of the mitochondrial in vitro studies we have to accept until in vivo methods will be available, if ever. In fact one of the major plagues in mitochondrial papers, is that many groups basically measure O2 consumption on screwed out mitochondria, because of poorly prepared samples!!
- -Page 9: found not clear
- -Personally don’t like »P«
- -ROX: not necessarily non mitochondrial respiration in absolute terms, but in the practice the ROX should be almost undectable if the instrument has been properly calibrated.
- EG: There are non-ETS enzymes catalyzing O2-consuming reactions.
- -it may be useful to mention how tricky it is to express fluxes by mass in weight g. The tissue composition in fact can change drastically between different diseases or individuals so that 1 g of dystrophic muscle will have half of its muscle mass replaced by fibrous tissue. Same is true for many other conditions, not to mention how drastically can differences in hydratation or even in the scale measurements (if very small pieces are used) of the tissue lead to errors.
- -not really clear how to normalize for mitochondrial mass: (a) stress the importance to measure the normalizer on the SAME prep introduced into the chamber, which can be sometime difficult or impossible. (b) I don’t think TOM20 can be used to normalize respirometric data (with which technique?) (c) mtDNA quantification has been shown to be a poor marker of mitochondrial mass, and is frequently altered in diseases indedndently from mass. Better use CS or caldiolipin. (d) I personally don’t agree on recommending using FCR as internal marker, it clearly changes between individuals and even in the preparation phase
- Of course I do not expect that everybody should agree, and open to discussion.
- As discussed in a previous workshop together, I believe this is a great initiative since so much confusion is in the general science community, especially since the recent resurrection of the interest for mitochondria from different specialists not previously involved in mitochondrial research. My very personal point of view is that the manuscript should be a bit restyled in order to focus on LESS but VERY clear concepts that can be easily digested by non specialists, in order to have a larger audience and impact, otherwise the paper will mainly talk to specialist, many of which are at least partly familiar with these informations. Please have a look how effectively these papers addressed methodological issues on cell death and autophagy. They are very easy to read and had huge impact. I would personally try to simplify the key concepts, cut energically overclassifications and formulae, limit historical citations, and possibly add a couple of experiments to illustrate the different fluxes. Limit abbreviations to a minimum. And more clearly explain what are OXPHOS and ETS, what it relies on, and what it means if 1 or both are decreased. Possibly with an illustrative trace. For the rest, with this first read, I have some other, more circumscribed comments:
- 2017-08-29 D Montaigne:
- As a whole the manuscript reads easily. I have just few comments/suggestions. Please note that I worked on the version 24’ / 2017-08-25.
- 1- would it be useful to evoke/include the beta-oxidation cycle in the general framework of Fig1A ?
- EG: We should do this in a follow-up part focussing on ETS pathways.
- 2- In Fig 1, H+out/O2 and H+in/»P ratios are presented as fixed ratios. My understanding is that these are the ratios expected while one NADH (and not one succinate) is oxydised in theory, e.g. not taking into account proton slipping. If I am correct, this should be precised in the fig legend ?
- EG edited the caption: H+out/O2 is the ratio of outward proton flux from the matrix space to catabolic O2 flux in the NADH-linked pathway. H+in/»P is the ratio of inward proton flux from the inter-membrane space to the endergonic flux of phosphorylation of ADP to ATP. Due to proton leak and slip these are not fixed stoichiometries.
- 3- at the bottom of page 10, « In isolated mammalian mitochondria ATP production catalysed by adenylate kinase…. » => what is the point of this sentence? particularly regarding mitochondrial exploration ?
- 4- Page 19, paragraph 3.1 : « The protonmotive force is high in the OXPHOS state …, elevated in the LEAK state… » I would change elevated into very high or maximum to make it clearer. - DONE.
- thanks again for letting me review this great manuscript.
- 2017-08-29 CM Palmeira:
- After all the suggestions that have been added, I just checked in the MitoEAGLE the last version, at the moment I have no additional comments. I think it is a great contribution to the field, and it would be great if the community will start to following the recommendations.
- 2017-08-28 Petit PX:
I agree with the almost final version of the text. I will follow the final position of all the partners but better BBA and Cell Metabolism if accepted. I find the text very pedagogic indeed.
- 2017-08-28 EG: Suggestions
- From 'Mitochondrial respiratory control: MitoEAGLE recommendations'
- To 'The protonmotive force and respiratory control: Building blocks of mitochondrial physiology'
- Figure 8 and Table 6: mte vs. mt
- 2017-08-27 Table 4 update by EG
- Protonmotive force, symbol ∆pH+ changed from previous symbol ∆pmt, for consistency of force and flow, FH+/e and IH+/e
- ∆pH+ = ∆Ψmt + ∆µH+ / F ; (Eq. 1)
- 2017-08-27 Morten Scheibye-Knudsen
- Regarding the journal, if you get a large consensus which includes a large contingency of authors I guess this could be of interest for a higher level journal because it could act as a reference point. Perhaps even something like Cell Metabolism. - I have attached a version with a some suggestions as you can see. It is obviously a great reference manuscript for mitochondrial investigations and I have only a few corrections/comments. I think the mitochondrial marker section is a bit simplistic and I would suggest including something about appropriate markers (for example use markers of beta-oxidation if you are investigating beta oxidation etc.).
- EG: Perhaps TOM20 is the better marker, if fatty acid oxidation (FAO) capacity is selectively stimulated relative to NADH- and succinate-linked pathway capacity, then enzymes involved in FAO are a good marker for FAO but a bad marker for mitochondrial content.
- Regarding the journal, if you get a large consensus which includes a large contingency of authors I guess this could be of interest for a higher level journal because it could act as a reference point. Perhaps even something like Cell Metabolism. - I have attached a version with a some suggestions as you can see. It is obviously a great reference manuscript for mitochondrial investigations and I have only a few corrections/comments. I think the mitochondrial marker section is a bit simplistic and I would suggest including something about appropriate markers (for example use markers of beta-oxidation if you are investigating beta oxidation etc.).
- 2017-08-26 Sophie Breton:
- First, I want to thank you for sharing the manuscript. Since I am more a "mitochondrial geneticist" than a mitochondrial physiologist, I found the manuscript on mitochondrial respiratory control very clear and well written, and it will be very helpful for my students (and also for myself actually)!! I have very few edits (attached is the manuscript).
- 2017-08-24 Journal: Int J Biochem Cell Biol, discussed at MITOEAGLE Barcelona 2017 and MiPschool Obergurgl 2017; page limit is surpassed.
- 2017-08-24 Garry Buettner
- I fully appreciate pushing the community to a common language and lab approaches so that data gathered can be useful far beyond the specific question of an experiment.
- 2017-08-24 David Fell:
- I've read through your draft and I think there is an issue that needs to be clarified, which is the distinction between 'responds to' and 'controlled by'. In MCA terms, this is equivalent to the difference between (flux) control coefficients and response coefficients. I could prepare a short document for you and your co-authors to explain my thinking on this issue. If you accept my points, the second step would be to work out how to alter the text without increasing its length. A question I have for you as well is: when you say 'respiratory control' are you exclusively concerned with control of oxygen consumption flux, or do you mean more generally it and other variables of the system, such as phosphorylation flux, or even PMF? If the latter, then there needs to be some clarification about that, in my view.
- EG: You raised two good and important points. I would welcome clarification between ‘responds to’ and ‘controlled by’, and editing critically the present version of the ms. will be a big improvement. Yes, ‘respiratory control’ should be clarified in a wider sense not restricted to oxygen flux, since we explicitely talk about states and rates. Again, clarification will help.
- I've read through your draft and I think there is an issue that needs to be clarified, which is the distinction between 'responds to' and 'controlled by'. In MCA terms, this is equivalent to the difference between (flux) control coefficients and response coefficients. I could prepare a short document for you and your co-authors to explain my thinking on this issue. If you accept my points, the second step would be to work out how to alter the text without increasing its length. A question I have for you as well is: when you say 'respiratory control' are you exclusively concerned with control of oxygen consumption flux, or do you mean more generally it and other variables of the system, such as phosphorylation flux, or even PMF? If the latter, then there needs to be some clarification about that, in my view.
- 2017-08-24 Graham R Scott:
- I think this is a very important project, and it will be fantastic to have this resource (and your future MitoEAGLE work) available to mitochondrial physiologists. I’m happy to add my name as a co-author if it is helpful. I don’t have much to add to the manuscript, its already very clear and I agree whole-heartedly with nearly all of what is written. I have a few editorial comments for you to consider below.
- Figure 1A: It might be simpler to show just one single arrow rather than multiple arrows between CI/CII and Q. I like the introduction of the term ">>P”, but when I first looked at Figure 1 I didn't understand what the ">>" represented because it hadn’t yet been defined, so consider defining it in the figure legend.
- EG: Yes, the definitions are added to the figure legend, where the multiple convergent electron transfer pathways are also explained.
- Figure 1B: The use of "-“ to reflect stoichiometry could be confusing to some readers, and it might be worth explaining it in the legend.
- Page 9: I find the term "evolutionary background" confusing. In this context, "evolutionary or acquired differences in the genetic basis of mitochondrial function (or dysfunction) between subjects" would be clearer, at least from my perspective.
- Figure 3: Should ATPase activity and the J<<P arrow be shown here? Could it be removed or somehow shown as lower than the rate of phosphorylation? Showing it as it is currently is relevant to intact cells, but less relevant to isolated mitochondria with low ATPase activity.
- EG: – But highly relevant for permeabilized cells and fibres.
- Page 16: I think a statement, either here or earlier, that explicitly distinguishes the terms uncoupled, noncoupled, and dyscoupled would be valuable. As written, the distinction is only made implicitly, but I think it would be valuable to new researchers to have the meanings spelled out clearly.
- Page 18, last two sentences: I think the point of these two sentences could be made more clearly.
- Page 35, “300 mitochondria per cell”: I think it would be better to express this idea by instead using the volume density of mitochondria per cell.
- I think this is a very important project, and it will be fantastic to have this resource (and your future MitoEAGLE work) available to mitochondrial physiologists. I’m happy to add my name as a co-author if it is helpful. I don’t have much to add to the manuscript, its already very clear and I agree whole-heartedly with nearly all of what is written. I have a few editorial comments for you to consider below.
- 2017-08-23 Version 22' (see Table 4):
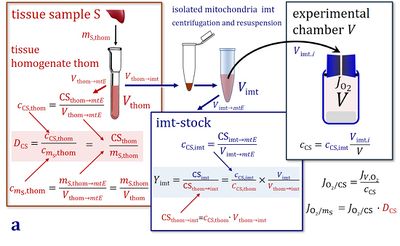
- Is there a general symbol available for amount of mitochondria, as estimated by a mitochondrial marker?
- Amount of mt-elements, mte (= quantity of mt-marker)
- Mitochondrial concentration in the instrumental chamber with volume V, Cmte=mte∙V-1
- Specific mitochondrial density in tissue X with tissue mass mX, Dmte=mte∙mX-1
- Mitochondrial content per cell, mtecell=mte∙Ncell-1 (where Ncell is the number of cells)
- 2017-08-22 Anibal E. Vercesi:
- I have carefully red version 20 and in my opinion is a fantastic peace of work. Up to this moment I did not have any recommendation for changes or improvements. I will follow the new versions. It is a honor for me to participate in this project.
- 2017-08-21 New Table 3 in Version 21
- 2017-08-19 Garry Buettner:
- Ad MITOEAGLE: Have all CAPS is a bit gaudy. It makes text look strange and uninviting. Because „ito“ are not „first“ letters, I suggest lower case.
- Erich: I agree. MITOEAGLE was originally used in our successful COST Action grant application, accoring to the COST rules to use capital letters for the project acronym. Since then I have seen several COST projects using small and capital letters for their acronym. MitoEAGLE looks better. Probably, we will need a Management Committee E-vote. Let's try MitoEAGLE for the manuscript, and decide on the general strategy.
- I would use L and g rather than mL and mg. Although l for liter is still in use, L is better and is the future. For example, the American Chem Soc Journals go with L as do many others. Elsevier is lagging, but becoming flexible.
- Erich: This makes sense, and we should get used to it. I added a conversion table, to facilitate harmonization.
- 2017-08-18 Erich edited some equations in Version 18:
- Protonmotive force, ∆pmt: The protonmotive force, ∆pmt,
- ∆pmt = ∆Ψmt + ∆µH+ / F ; (Eq. 1)
- is composed of an electric part, ∆Ψmt, which is the difference of charge (electrical potential difference) across the inner mitochondrial membrane, and a chemical part, ∆µH+/F, which stems from the difference of pH (chemical potential difference) across the inner mitochondrial membrane and incorporates the Faraday constant, F. In other words, the protonmotive force is expressed as the sum of two terms, with somewhat complicated symbols in Eq. 1, which can be more easily explained as isomorphic partial protonmotive forces.
- Electrical, el: Fe,el = ∆Ψmt is the electrical pat of the protonmotive force expressed in units joules per coulomb, i.e., volt [V=J/C], and defined as partial Gibbs energy change per motive elementary charge of protons, e [C]. Fn,el = ∆Ψmt∙F is the electrical force expressed in units joules per mole [V=J/mol], and defined as partial Gibbs energy change per motive amount of charge, n [mol].
- Chemical, diffusion or dislocation, d: Fn,d = ∆µH+ is the corresponding chemical part of the protonmotive force expressed in units joules per mole [J/mol], and defined as partial Gibbs energy change per motive amount of protons, n [mol]. Fe,d = ∆µH+/F is the chemical force expressed in units joules per coulomb, i.e., volt [V=J/C], and defined as partial Gibbs energy change per motive amount of protons expressed in units of electric charge, e [C].
- Faraday constant, F: The Faraday constant is the product of the elementary charge and the Avogadro (or Loschmidt) constant, F = e∙NA [C/mol], and yields the conversion between protonmotive force, Fe = ∆pmt [J/C], expressed per motive charge, e [C], and protonmotive force or chemiosmotic potential difference, Fn = ∆pmt∙F [J/mol], expressed per motive amount of substance, n [mol],
- Fn = Fe ∙ F ; (Eq. 2.1)
- Fe = Fe,el + Fe,d = ∆Ψmt + ∆µH+/F ; e-isomorph [J/C=V] (Eq. 2.2)
- Fn = Fn,el + Fn,d = ∆Ψmt∙F + ∆µH+ ; n-isomorph [J/mol] (Eq. 2.3)
- Protonmotive means that protons are moved across the mitochondrial membrane at constant force, and the direction of the translocation is defined in Fig. 2 as H+in → H+out,
- Fn,d = ∆µH+ = -ln(10)∙RT∙∆pHmt ; (Eq. 3)
- where RT is the gas constant times absolute temperature. ln(10)∙RT = 5.708 and 5.938 kJ∙mol-1 at 25 and 37 °C, respectively. ln(10)∙RT/F = 59.16 and 61.54 mV at 25 and 37 °C, respectively. For a ∆pH of 1 unit, the chemical force (Eq. 3) changes by 6 kJ∙mol-1 and the protonmotive force (Eq. 2.2) changes by 0.06 V.
- Since F equals 96.5 (kJ∙mol-1)/V, a membrane potential difference of -0.2 V (Eq. 2.2) equals a chemiosmotic potential difference, Fn, of 19 kJ∙mol-1 H+outSubscript text (Eq. 2.3). Considering a driving force of -470 kJ∙mol-1 O2 for oxidation, the thermodynamic limit of the H+out/O2 ratio is reached at a value of 470/19 = 24, compared to a mechanistic stoichiometry of 20 (H+out/O=10).
- Protonmotive force, ∆pmt: The protonmotive force, ∆pmt,
- 2017-08-17 Perspective added in Version 17 (EG)
- To provide an overall perspective of mitochondrial physiology we may link cellular bioenergetics to systemic human respiratory activity, without yet addressing cell- and tissue-specific mitochondrial function. A routine O2 flow of 234 µmol∙s-1 per individual or flux of 3.3 nmol∙s-1∙g-1 body mass corresponds to -110 W catabolic energy flow at a body mass of 70 kg and -470 kJ/mol O2. Considering a cell count of 514∙106 cells per g tissue mass and an estimate of 300 mitochondria per cell (Ahluwalia 2017), the average oxygen flow per million cells at Jm,O2peak of 45 nmol·s-1·g-1 (60 ml O2·min-1·kg-1) is 88 pmol∙s-1∙10-6 cells, which compares well with OXPHOS capacity of human fibroblasts (not ETS but the lower OXPHOS capacity is used as a reference; Gnaiger 2014). We can describe our body as the sum of 36∙1012 cells (36 trillion cells). Mitochondrial fitness of our 11∙1015 mitochondria (11 quadrillion mt) is indicated if O2 flow of 0.02 pmol∙s-1∙10-6 mt at rest can be activated to 0.3 pmol∙s-1∙10-6 mt at high ergometric performance.
- 2018-08-16 EG - added Fig. 6 and text in Version 16:
- Fig. 6 summarizes the three coupling states, ETS, LEAK and OXPHOS, and puts them into a schematic context with the corresponding respiratory rates, abbreviated as E, L and P, respectively. This clarifies that E may exceed or be equal to P, but E cannot theoretically be lower than P. E<P must be discounted as an artefact, which may be caused experimentally by (i) using high and inhibitory uncoupler concentrations (Gnaiger 2008), (ii) non-saturating [ADP] or [Pi] (State 3), (iii) high oligomycin concentrations applied for measurement of L before titrations of uncoupler, when oligomycin exerts an inhibitory effect on E, or (iv) loss of oxidative capacity during the time course of the respirometric assay with E measured subsequently to P (Gnaiger 2014). On the other hand, E>P is observed in many types of mitochondria and depends on (i) the excess ETS capacity pushing the phosphorylation system (Fig. 1B) to the limit of its capacity of utilizing ∆pmt, (ii) the pathway control state with single or multiple electron input into the Q-junction and involvement of three or less coupling sites determining the H+out/O2 coupling stoichiometry (Fig. 2A), and (iii) the biochemical coupling efficiency expressed as (E-L)/E, since any increase of L causes an increase of P upwards to the limit of E. The excess E-P capacity, ExP=E-P, therefore, provides a sensitive diagnostic indicator of specific injuries of the phosphorylation system, when E remains constant but P declines relative to controls (Fig. 6). Substrate cocktails supporting simultaneous convergent electron transfer to the Q-junction for reconstitution of TCA cycle function stimulate ETS capacity, and consequently increase the sensitivity of the ExP assay.
- ... In general, it is inappropriate to use the term ATP production for the difference of oxygen consumption measured in states P and L. The difference P-L is the upper limit of the part of OXPHOS capacity which is free (corrected for LEAK respiration) and is fully coupled to phosphorylation with a maximum mechanistic stoichoimetry, ≈P = P-L (Fig. 6).
- 2018-08-16 Nicoleta Moisoi:
- I think that it is integrating well the 'State 1-5 - nomenclature' with the 'new terminology'. In 2.2 or in the final paragraph: A schematic view similar with the one from the previous versions of the manuscript (April 2017, on which I have sent feedback at the time) (see above) may still be useful for readers that are new to the field of mitochondria physiology, but are starting to employ such measurements in their work.
- 2018-08-16 David G Nicholls:
- Thank you for the invitation to contribute to your review. However, there is too great a distance between your in-depth approach and the minimalistic ‘need-to-know’ proton circuitry that I have promulgated for the past 40 years or so, and so I must decline. Good luck with the review.
- 2018-08-16 Dorit Ben-Shachar:
- I went through your interesting review which for me also enlightened new aspects on the mitochondrial chemophysical properties. I completely agree with the need to establish a common language and standard criteria in the field of mitochondria, so that different studies can be compared and abnormalities associated with diseases will be defined and maybe used in the future as biomarkers. I introduced several changes and asked for several clarifications by comments and notes in the text. As I am not an expert in fluxes and flows, forces and rates my input to these parts is limited. I do suggest to include a short paragraph on the crosstalk between mitochondria and the cells via Ca, and other signaling pathways which may affect mitochondrial activity in the conclusion part. Although it is beyond the scope of the review it puts it in a wider physiological context.
- 2018-08-16 David Harrison:
- Here are my minor comments on the manuscript. .., but I hope that you find them useful. As I comment in the abstract, I am not sure to what extent prior knowledge of the existing terminology can be assumed. However, it is clearly aimed at a readership already active in the field.
- 2018-08-16 Luís Crisóstomo:
- As for our concern, this version can get our approval. However, we think the text can be improved in the aspect of linguistic coherence. There are sentences that seem to not connect well, maybe because they were written by different authors. For instance, the last paragraph of the conclusion, seems to be more suitable for an introduction, rather than for a conclusion. The content of the text, in our opinion, is very good, very clear and useful for an harmonization of terms. The major terminology issues about mitochondrial states are addressed, and the text sets the paradigm for the upcoming parts. As mentioned in the ms, there will be the need of a "part 2", totally dedicated to mitochondrial enzyme nomenclature. We hope we are able to contribute with more critic comments on that subject, as it is more related to our expertise thus far, and a more active role on the endeavor.
- 2018-08-16 Leo Sazanov: Yes, will be happy to contribute - I think some changes to Fig. 1 are needed.
- 2018-08-16 Yau-Huei Wei: I have completed reading and revising this manuscript (attached). I appreciate your enthusiasm and great effort in preparing this manuscript.
- 2018-08-15 Jenny Collins:
- I added in a few comments on the attached PDF. I think it looks great and I’m glad to have been a part of the process. 'This bubble feels a bit disjointed from the rest of the paper. I'm not sure that it's necessary since it's never referred back to.'
- 2018-08-15 Mike Davis:
- I appreciate the opportunity to contribute to this joint review. Please give me a few days to examine the most recent version – if I feel that I understand the material well enough to comment and/or approve, I will be happy to do so and join the co-author list. One comment that I do have based on the preliminary reading that I did over my morning coffee is that for someone like me who views all of this as more of a means to an end (the end being understanding physiology and pathophysiology on a more macroscopic level) instead of an end unto itself, I find that when I am struggling to understand something it is because it lacks a context for me – either from a methodological standpoint (if you add X and Y to the reaction mixture, the results represent Z) or a physiological standpoint (adding X and Y and getting Z represents these in vivo conditions). Context is not necessary for someone like yourself and many of the co-authors who live in the world of mitochondrial physiology, but in order to gain a broader acceptance of the revised terminology and concepts, that additional context may be necessary.
- 2017-08-15 Pablo Garcia Roves:
- Find attached my comments after reading the manuscript. There is a lot of work behind the content of this manuscript. Some parts will be more difficult to follow for scientists new to the field of mitochondrial physiology but the information supply is correct, careful and hopefully an starting point to start unifying terminology.
File:OXPHOS system.jpg
Fig. 1. The mitochondrial respiratory system. In oxidative phosphorylation the electron transfer system (A) is coupled to the phosphorylation system (B). See Eqs. 4 and 5 for further explanation. Modified from (A) Lemieux et al (2017) and (B) Gnaiger (2014).
- 2017-08-15 New Fig. 1 in Version 15 (EG).
- 2017-08-15 Tony Moore:
- What happens when the ETS is branched at the oxidase level - need to define what "uncoupled" is!
- Added text: "Such uncoupling is different from switching to mitochondrial pathways which involve less than three coupling sites with electron entry into the Q-junction bypassing Complex I (Fig. 1; including a bypass of CIV by alternative oxidases, not shown). This may be considered as a switch of gears (stoichiometry) rather than uncoupling (loosening the stoichiometry)." EG
- the symbol »P: Ambiguous and likely to cause confusion! In my view you need to also define that you have changed this to »P/O2 and not O indicating you are referring to the full 4 electron reduction of oxygen!
- Added to the text!
- How are you going to define the ROX state in tissues that have had additional oxidase added i.e. AOX following Gene therapy.
- AOX has to be inhibited, too, to measure ROX, since AOX is part of the (genetically modified) electron transfer system. See original text: "ROX is measured either in the absence of fuel substrates or after blocking the electron supply to cytochrome c oxidase and alternative oxidases."
- FO2 versus F
- Added after Eq. 1: ".. and a chemical part incorporating the Faraday constant"
- 2017-08-15 Beata Velika: I have read joint review, it is very nice manuscript, and very useful for teaching. I really like the chapter Normalization: flows and fluxes, its very good expalined.
- 2017-08-14 EG added to Version 14 many suggestions of GC Brown, and
- Coupled versus bound processes: Since the chemiosmotic theory explains the mechanism of coupling in OXPHOS, it may be interesting to ask if the electrical and chemical parts of proton translocation are coupled processes. This is not the case according to the definition of coupling given above (in the manuscript). It is not possible to physically uncouple the electrical and chemical processes, which are only theoretically partitioned as electrical and chemical components (Eq. 1) and can be measured separately. If partial processes (fluxes, forces) are non-separable, i.e. cannot be uncoupled, then these are not coupled but are defined as bound processes. The electrical and chemical part of Eq. 1 are tightly bound partial forces of the protonmotive force.
- 2017-08-14 Guy Brown:
- "I have very fond memories of the MiP summer schools.I can’t help much with this, but I enclose some thoughts.In general, the purpose is a bit unclear, and it rambles over a variety of areas, without clear definitions, recommendations or focus. If I were doing it, I would half the length and sharpen the focus, and make the definitions and recommendations crystal clear. But I realise that doing this within a large consortium is not easy!"
- Response by EG: As always, I fully appreciate your sharp and to-the-point level of discussion. Like me, many of us consider you as a teacher. Therefore, I respond with full appreciation that you ‘help us with this’. And yes: the large consortium has by now a history of dedicated meetings with good discussions, resulting in an evolutionary approach that needs to be appreciated without the claim of full-power selective optimization. I thank you so much for your detailed feedback. Many suggestions that you made are implemented in the new version. In addition my comments are summarized here:
- Abstact: This was written before the group-dynamics changed the focus of the ms. With more and more questions on ‘clarification’, ‘definition of terms’, the attempt to summarize some simple recommendations shifted to ‘educational’. So far, more ‘explanatory’ was the result, without reassertion if this was also more ‘educational’. We need the help of experts in science writing who are firm with the basic concepts.
- I agree (without necessarily speaking of other contributors) that we should reduce ‘crazy’ recommendations on abbreviations. It is a cancerous phenomenon of scientific literature, and we better stay away of too much of the same. imt etc are not even used in later sections, thus we can get rid of it.
- I suggest an exception of the above agreement toavoid abbreviations: mt. Would you suggest that we recommend to use ‘mitochondrial DNA’ and skip mtDNA?
- tr: consider spelling this out in all following symbols.
- Section 2.2. “You need a section here, outlining why the classical nomenclature is not sufficient. This is important!“ - Thanks, there should be more detail in the first introductory sentences.
- LEAK state - thanks, would you agree on this: “A state of mitochondrial respiration when oxygen flux is maintained at saturating levels of oxygen and respiratory substrates, and zero ATP-turnover without addition of any experimental uncoupler, as an estimate of the maximal proton leak rate.”
- Your excellent comments on detail have all been incorporated in Version 14.
- "∆pmt - not clear why you are using mt here." Protonmotive force could be used for any membrane, but in the context of this manuscript, confusion is very unlikely." – But think of all the literature not taking into account the plasma membrane potential. - Later versions: "∆pmt changed to "∆pH+
- Response by EG: As always, I fully appreciate your sharp and to-the-point level of discussion. Like me, many of us consider you as a teacher. Therefore, I respond with full appreciation that you ‘help us with this’. And yes: the large consortium has by now a history of dedicated meetings with good discussions, resulting in an evolutionary approach that needs to be appreciated without the claim of full-power selective optimization. I thank you so much for your detailed feedback. Many suggestions that you made are implemented in the new version. In addition my comments are summarized here:
- “The protonmotive force is maximum in the LEAK state” - I agree with your comment – is “elevated” better?
- "Forces and flows in physics and irreversible thermodynamics: Why?" – Can we ignore IUPAC recommendations?
- 2017-08-14 Masashi Tanaka (response by EG):
- On Fig. 1A: Perhaps we should simplify it for the present purpose, since the chapter on ‘pathway control’ has been shifted away form the present Part 1 to another manuscript (Part 2), where these issues should be discussed and controversies resolved in full detail.
- Intact cell respiration was also shifted to another future manuscript.
- I added ‘intermembrane space’ to Fig. 1B.
- V[dot]O2max: I fully agree, I just do not know how to add the dot in Microsoft Word. In the old style, it was simple to just put such a dot on paper.
- Definition of V: It is unfortunate that V[dot]O2max has been introduced, but we cannot expect to change the sport science symbols (they should change from volume to amount of substance for metabolic oxygen consumption). And we cannot change V. Therefore V[dot] needs to be distinguished from the other definitions of V. I added: “.. whereas maximum mass-specific oxygen flux, V[dot]O2max or V[dot]O2peak, is constant across a large range of individual body mass (Weibel and Hoppeler 2005). V[dot]O2peak of human endurance athletes is 60 up to 80 ml O2·min-1·kg-1 body mass, converted to Jm,O2peak of 45 to 60 nmol·s-1·g-1 (Gnaiger 2014).”
- 2017-08-13 Brian Irving: The review is coming together nicely. Much more advanced than a couple weeks ago. Great job. I added a few comments.
- 2017-08-12 EG added to Version 12
- Proton leak: Proton leak is the process in which protons are translocated across the inner mt-membrane in the direction of the downhill protonmotive force without coupling to phosphorylation. The proton leak flux depends on ∆pmt and is a property of the inner mt-membrane. Proton slip: Proton slip is the process in which protons are only partially translocated by a proton pump and slip back to the original compartment. The proton slip is a property of the proton pump and depends on the turnover rate of the proton pump. - (added by EG)
- 2017-08-12 Feedback and suggestions on Version 10 by A Molina.
- I managed to set aside some time to work on the manuscript. My suggested revisions and comments are embedded in the document using the “track changes” mode and margin comments.
- On »P (»P/O ratio) and «P: Can a stronger statement be made here? I like the suggested symbols.
- On ADP concentration: It may be useful to discuss the potential confusion between high ADP and saturating ADP. The arbitrariness of some commonly used protocols is a problem in the field, particularly when using plate-based systems for measuring respiration.
- I managed to set aside some time to work on the manuscript. My suggested revisions and comments are embedded in the document using the “track changes” mode and margin comments.
- 2017-08-11 Feedback and suggestions on Version 9 by J Iglesias-Gonzalez.
- 2017-08-11 HK Lee:The chapter on protonmotive force is difficult to read.
- Thank you for your generous invitation to become an author of this historical paper. This is a great honor to me. I have some comments to make;
- I wish you would elaborate more on the respiratory 'control', I always wondered how and why this term is adopted, instead of other terms, such as mitochondrial functional characteristics or functional anatomy.
- Addition to the MS by EG: Control and regulation: The terms metabolic control and regulation are frequently used synonymously, but are distinguished in metabolic control analysis (Fell 1997). Respiratory control may be exerted by (1) ATP demand (Fig. 2), (2) fuel substrate, pathway competition and oxygen availability (starvation and hypoxia), (3) the protonmotive force, redox states, flux-force relationships, coupling and efficiency, (4) mitochondrial enzyme activities and allosteric regulation by adenylates, phosphorylation of regulatory enzymes, Ca2+ and other ions including pH, (5) inhibitors (e.g. NO or intermediary metabolites, such as oxaloacetate), (6) enzyme content, concentrations of cofactors and conserved moieties) such as adenylates, NADH/NAD+, coenzyme Q, cytochrome c); (7) metabolic channeling by supercomplexes, (8) mitochondrial density and morphology (fission and fusion), (9) hormone levels, gender, life style (influencing all control mechanisms listed before), and (10) genetic or acquired diseases causing mitochondrial dysfunction (for reviews see Brown 1992; Gnaiger 1993a, 2009; 2014; Morrow et al 2017).
- I was really happy to find a section "Size-specific quantities", where you wrote "The well-established scaling law in respiratory physiology reveals a strong interaction of oxygen consumption and body mass by the fact that mass-specific basal metabolic rate (oxygen flux) does not increase proportionally and linearly with body mass, whereas maximum mass-specific oxygen flux, VO2max, is constant across a large range of body mass (Weibel and Hoppeler 2005)." However, I understand the mass-specific (basal) metabolic rate (oxygen flux) decrease proportionally if not linearly with body mass.
- I found the discussion on the protonmotive force very challenging. I wonder if you could make it simpler for more wide audiences.
- Thank you for your generous invitation to become an author of this historical paper. This is a great honor to me. I have some comments to make;
- 2017-08-10 Version 10: Integration of suggestions and corrections by T Komlodi and A Meszaros.
- Phosphorylation, »P: Although phosphorylation in the context of OXPHOS is clearly defined as phosphorylation of ADP to ATP, potentially involving substrate-level phosphorylation as part of the tricarboxylic acid cycle (succinyl-CoA ligase), in the matrix (phosphoenylpyruvate carboxykinase) and in the cytosol (pyruvate kinase, phosphoglycerate kinase). ADP is formed in the adenylate kinase reaction, 2 ADP <--> ATP + AMP. In isolated mitochondria high adenylate kinase related ATP production can be detected in the presence of ADP and without respiratory substrates (Komlódi and Tretter 2017). On the other hand, the term phosphorylation is used in the general literature in many different contexts (phosphorylation of enzymes, etc.). This justifies consideration of a symbol more discriminative than P as used in the P/O ratio (phosphate to atomic oxygen ratio), where P indicates phosphorylation of ADP to ATP or GDP to GTP. We propose the symbol »P for the energetic uphill direction of phosphorylation coupled to catabolic reactions, and likewise the symbol «P for the corresponding downhill reaction (Fig. 2).
- Coupling and efficiency: In an energy transformation, tr, coupling occurs between processes, if a coupling mechanism allows work to be performed on the endergonic or uphill output process (work per unit time is power; dW/dt [J/s] = Pout [W]; with a positive partial Gibbs energy change) driven by the exergonic or downhill input process (with a negative partial Gibbs energy change). At the limit of maximum efficiency of a completely coupled system, the (negative) input power equals the (positive) output power, such that the total power equals zero at an efficiency of 1. If the coupling mechanism is disengaged, the output process becomes independent of the input, and both proceed in their downhill direction (Fig. 2). - (added by EG)
- Extensive quantities: An extensive quantity increases proportional with system size. The magnitude of an extensive quantity is completely additive for non-interacting subsystems, such as mass or flow expressed per defined system. The magnitude of these quantities depends on the extent or size of the system (Cohen et al 2008).
- Size-specific quantities: ‘The adjective specific before the name of an extensive quantity is often used to mean divided by mass’ (Cohen et al 2008). A mass-specific quantity (e.g. mass-specific flux is flow divided by mass of the system) is independent of the extent of non-interacting homogenous subsystems. Tissue specific quantities are of fundamental interest in comparative mitochondrial physiology, where specific refers to the type rather than mass of the tissue. The term specific, therefore, must be clarified further, such that tissue mass-specific (e.g. muscle mass-specific) quantities are defined. - (added by EG)
- 2017-08-08 to 09: Updated versions by EG, with Sections 3.2. Normalization: flows and fluxes and 3.3. Conversion: oxygen, protons, ATP
- Forces and flows in physics and irreversible thermodynamics: According to definition in physics, a potential difference and as such the protonmotive force, ∆pmt, is not a force (Cohen et al 2008). The fundamental forces of physics are distinguished from motive forces (e.g. ∆pmt) of statistical and irreversible thermodynamics. Complementary to the attempt towards unification of fundamental forces defined in physics, the concepts of Nobel laureates Lars Onsager, Erwin Schrödinger, Ilya Prigogine and Peter Mitchell (even if expressed in apparently unrelated terms) unite the diversity of ‘isomorphic’ flow-force relationships, the product of which links to the dissipation function and Second Law of thermodynamics (Prigogine 1967; Schrödinger 1944). A motive force is the change of potentially available or ‘free’ energy (exergy) per isomorphic motive unit (force=exergy/motive unit; in integral form, this definition takes care of isothermal and non-isothermal processes). A potential difference is, in the framework of flow-force relationships, an isomorphic force, Ftr, involved in an exergy transformation, defined as the partial derivative of Gibbs energy, ∂trG, per advancement, dtrξ, of the transformation, tr (the isomorphic motive unit in the transformation): Ftr = ∂trG/dtrξ (Gnaiger 1993a,b). This formal generalization represents an appreciation of the conceptual beauty of Peter Mitchel’s innovation of the protonmotive force against the background of the established paradigm of the electromotive force (emf) defined at the limit of zero current (Cohen et al 2008).
- Coupling, efficiency and power: In energetics (ergodynamics) coupling is defined as an exergy transformation fuelled by an exergonic (downhill) input process driving the advancement of an endergonic (uphill) output process. The (negative) output/input power ratio is the efficiency of a coupled energy transformation. Power, Ptr = ∂trG/dt [W=J∙s-1], is closely linked to the dissipation function (Prigogine 1967) and is the product of flow, Itr=dtrξ∙dt-1 [xtr∙s-1] times generalized force, Ftr = ∂trG/∂trξ [J∙xtr-1] (Gnaiger 1993b).
- 2017-08-02 Updated ms summarizing WG1 input by J Iglesias-Gonzalez.
- 2017-08-01 Javier Iglesias-Gonzalez:
- I’ve just finished with the last version (9) of the paper. I really like how it looks like now and I have let some of our PhD students to read (as a test for a beginner) and she enjoyed a lot the paper. I added very few suggestions which I attached in the word document.
- 2017-07-29 Version 3 on website: http://www.mitoeagle.org/index.php/Mitochondrial_respiratory_control:_MITOEAGLE_recommendations_1
- 2017-07-28 to 29: Obergurgl MITOEAGLE Workshop: WG1 – group discussion and edits based on printed Version 2.
- 2017-04-21 Petit PX:
- I find the text clearly exposed even if the publicity for Mitoeagle is to much pronounced (but that is a choice).
- Concerning:
- State 1: depending on the fact that mitochondrial extract of isolated mitochondria is crude or purified (for exemple on Percoll gradients or sucrose gradients), the isolated mitochondria have still somes endogenous substrats or not that makes a pulse at the start during their early dissipation. Could this be taken into consideration or being indiscted for the users.
- State 4: I do not find this very well writen since after the addition of ADP (state 3) the mitochondrial membrane potential drop and reacquire its high value (at the state 4) when all the ADP has been transformed in ATP has been used? am I wrong. So this shoul be writen clearly.
- Why escaping to state about CR (respiratory control) and ADP/O measurements and definitions?
- Again, I perfectly understand the reference to the bioblast link by since we do not live outside teh real world the reference are usually referred to pubmed also... so we should refer to both...
- For te publication, the suggestion is nice but by tradition our journals are more....
- BBA general subjects or bioenergetic: 6554 occurrences
- FEBS J: 2911 FEBS letters 2563 (so 1+2 = 5474 Occurrences)
- Journal of Biological Chemistry: 7730 occurrences
- My personnal preference will go To BBA general subjects... But should be decided in commun
- 2017-04-18 Version 1 circulated to MITOEAGLE by E Gnaiger.
- 2017 Mar 21-23: Barcelona MITOEAGLE Workshop: WG1 – presentations and group discussions.
Position statements
- I agree with you and the purpose and philosophy that the paper pursues.