Difference between revisions of "User talk:Tindle Lisa/My sandbox Support page v2"
From Bioblast
Tindle Lisa (talk | contribs) |
Tindle Lisa (talk | contribs) m (Tindle Lisa moved page User talk:Tindle Lisa/My sandbox CureMILS to User talk:Tindle Lisa/My sandbox Support page v2 without leaving a redirect) |
||
| (12 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
{{Template:OROBOROS support page name}} | |||
<big> | :::::::::::: <big>''Currently Asked Questions - »'''[[O2k-Open Support alert]]'''« </big> | ||
[[File: | <!-- TODAY'S FEATURED CONTENT --> | ||
{| role="presentation" id="mp-upper" style="width: 100%; margin-top:4px; border-spacing: 0px;" | |||
<!-- 1ST BOX | CURRENT WEEK AGENDA - LEFT --> | |||
| id="mp-left" class="MainPageBG" style="width:75%; border:1px solid #99b4ed; padding:0; background:#f0f5ff; vertical-align:top; color:#000;"| | |||
<div id="mp-tfa-h2" style="margin:0.5em; background:#2856ba; font-family:inherit; font-size:120%; font-weight:bold; border:1px solid #a3bfb1; color:#ffffff; padding:0.2em 0.4em;"></div> | |||
<div id="mp-tfa" style="padding:0.1em 0.6em;"> | |||
__TOC__ | |||
== O2k-Open Support resources == | |||
[[File:Expand.png|right|45px |Click to expand or collaps]] | |||
<div class="toccolours mw-collapsible mw-collapsed"> | |||
'''Startup information''' | |||
<div class="mw-collapsible-content"> | |||
:::: Each O2k-Package includes a remote installation and startup support session with an Oroboros expert, which can be scheduled during any time within the O2k-Warranty period (max. 2 years) | |||
:::::» '''[https://www.oroboros.at/index.php/support-session-request-form/ Register for your session]''' | |||
:::::» [[Installation and startup support session self-study material]] | |||
:::: Upon special request and subject to an additional fee, Oroboros Instruments offers a start-up visit by a scientific application specialist. | |||
:::::» [[O2k on-site training]] | |||
:::: Additional reading | |||
:::::» [[MiPNet20.04 O2k-checklist| Getting started with an O2k-experiment]] | |||
:::::» O2k-Workshop self-study material: [https://wiki.oroboros.at/index.php/Virtual_O2k-Workshop_self-study_material#O2k-Basic O2k-Basic] | |||
</div> | |||
</div> | |||
[[File:Expand.png|right|45px |Click to expand or collaps]] | |||
<div class="toccolours mw-collapsible mw-collapsed"> | |||
'''Quality control''' | |||
<div class="mw-collapsible-content"> | |||
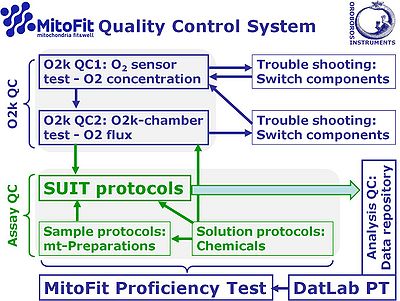
[[File:MitoFit-QCS.jpg|right|400px|MitoFit-QCS]] | |||
:::: Many commonly raised issues of O2k-technical support are solved in the newest version of [[DatLab]]. | |||
:::: The '''O<sub>2</sub> sensor and O2k chamber tests''' are important to eliminate instrumental artefacts (instrumental quality control, QC), distinguished from assay QC addressing problems of [[solution protocols]] (preparation of chemicals), [[sample]] preparation protocols (such as [[mitochondrial preparations]]), and [[SUIT protocols]]. | |||
::::# Clean O2k-chambers and contamination-free incubation medium are required. | |||
::::# Analysis of DatLab files recorded in instrumental QC tests is essential. | |||
::::# Biological experiments are not suitable for trouble shooting. | |||
:::: '''1. O<sub>2</sub> sensor test''' | |||
:::: The [[OroboPOS|OroboPOS]] (polarographic oxygen sensor) requires regular service at intervals - these may be more than 1 year. SOPs are available to determine if a sensor service is required at an earlier or later date: | |||
::::» O<sub>2</sub> sensor test: [[Oxygen_sensor_test |'''QC1: Oxygen sensor test''']] | |||
:::: '''2. O2k-chamber test''' | |||
:::: If all quality control criteria of the O<sub>2</sub> sensor test are met, the operator can be assured that the quality of the sensor signal is acceptable. Next, the quality of the O2k-chamber assembly has to be tested, described in detail: | |||
::::» O2k-chamber test: [[Oxygen flux - instrumental background |'''QC2: Instrumental O<sub>2</sub> background test''']] | |||
</div> | |||
</div> | |||
[[File:Expand.png|right|45px |Click to expand or collaps]] | |||
<div class="toccolours mw-collapsible mw-collapsed"> | |||
'''Advanced topics''' | |||
<div class="mw-collapsible-content"> | |||
::» O2k-Workshop self-study material: [https://wiki.oroboros.at/index.php/Virtual_O2k-Workshop_self-study_material#O2k-Advanced O2k-Applications: Simultaneous determination of O2 and H2O2 fluxes] | |||
:: You may also find the answers to more advanced topics using the navigation menu above or simply by using the Bioblast search function.: | |||
{| | |||
| | |||
| | |||
| | |||
| [[Image:O2k-Hardware icon.jpg|50px|link=https://wiki.oroboros.at/index.php/MitoPedia:_O2k_hardware |MitoPedia: O2k hardware]] | |||
| | |||
| | |||
| [[Image:DatLab icon.jpg|50px|link=https://wiki.oroboros.at/index.php/MitoPedia:_DatLab |MitoPedia: DatLab]] | |||
| | |||
| | |||
| [[Image:SUIT icon.jpg|50px|link=https://wiki.oroboros.at/index.php/MitoPedia:_SUIT |MitoPedia: SUIT]] | |||
| | |||
| | |||
| [[Image:O2k-Protocols.jpg|50px|link=http://wiki.oroboros.at/index.php/O2k-Procedures |O2k-Procedures]] | |||
| | |||
| | |||
| [[Image:O2k-Manual.jpg|50px|link=http://wiki.oroboros.at/index.php/O2k-Manual|O2k-Manual]] | |||
| | |||
| | |||
| [[Image:PlayVideo.jpg|50px|link=http://wiki.oroboros.at/index.php/O2k-Videosupport |O2k-Videosupport]] | |||
| | |||
| | |||
| [[Image:O2k-GlossaryFaq icon.jpg|50px|link=https://wiki.oroboros.at/index.php/MitoPedia:_O2k-Open_Support |MitoPedia: O2k-Open Support]] | |||
| | |||
| | |||
|} | |||
::::::# '''MitoPedia: O2k''': find information quickly with MitoPedia, with lists of key words and links to relevant pages. | |||
::::::## [[MitoPedia: O2k hardware]] | |||
::::::## [[MitoPedia: DatLab]] | |||
::::::## [[MitoPedia:_SUIT ]] | |||
::::::# [[O2k-Procedures]]: explain various applications of the O2k with O2k-Demo experiments | |||
::::::# [[O2k-Manual]]: topic specific manuals with step by step help | |||
::::::# [[O2k-Videosupport]]: provides valuable assistance, complementary to the O2k-Manual | |||
::::::# [[MitoPedia: O2k-Open Support|O2k-Glossary]]: full list of key words and links to relevant pages. | |||
</div> | |||
</div> | |||
== Ask an O2k-Expert and the O2k-Open Support agreement == | |||
:::: We give you the tools to be self-sufficient, but sometimes you need expert advice or help solving a technical problem. The Oroboros experts will be happy to help in using our O2k-Open Support system. » [https://www.oroboros.at/index.php/o2k-technical-support '''Contact Support'''] | |||
::::» [[O2k-Open Support agreement]] | |||
::::» [[O2k repair]] | |||
[[File:O2k-Network.png|left|100px|O2k-Network|link=O2k-Network]] | |||
== The O2k-Network: An additional resource == | |||
:::: The Oroboros O2k-Network serves to connect and support: Contact for advice on applications of the O2k and high-resolution respirometry - the global network in mitochondrial physiology. | |||
::::: '''»''' The O2k-Network Labs: [[O2k-Network|» O2k-Network]] | |||
::::: '''»''' [[O2k-Network_discussion_forum|O2k-Network discussion forum]] | |||
:::: '''»''' [[Oroboros Laboratories|Oroboros O2k-Laboratory]] - open for innovation and cooperation. | |||
:::: '''»''' [[Oroboros_Science_Scholarship|Oroboros Science Scholarships]] - are offered for focused research projects on mitochondrial physiology and high-resolution respirometry | |||
[[Category:O2k-Open Support]] | |||
<br /> | |||
</div> | |||
= | | style="border:1px solid transparent;"| | ||
=== | <!-- 2ND BOX | IN FOCUS - RIGHT --> | ||
:::: | | id="mp-right" class="MainPageBG" style="width:25%; border:1px solid #cedff2; padding:0; background:#f5faff; vertical-align:top;"| | ||
<div id="mp-tfa-h2" style="margin:0.5em; background:#2856ba; font-family:inherit; font-size:120%; font-weight:bold; border:1px solid #a3bfb1; color:#ffffff; padding:0.2em 0.1em;"></div> | |||
:::: | |||
== | <div id="mp-itn" style="padding:0.1em 0.6em;"> | ||
[[File:O2k-support system.jpg|center|350px|Oroboros support system]]</div> | |||
< | <!-- NEXTGEN-O2K BANNER --> | ||
< | <div style="margin:0.5em; background:#cedff2; font-family:inherit; font-size:120%; font-weight:bold; border:1px solid #a3b0bf; color:#000; padding:0.2em 0.1em;">Funding acknowledgments</div> | ||
< | <div id="mp-itn" style="padding:0.1em 0.1em;">[[File:Template NextGen-O2k.jpg|center|350px|NextGen-O2k]]</div> | ||
|} | |||
<br /> | |||
</div> | |||
Latest revision as of 13:59, 29 July 2021
 |
User talk:Tindle Lisa/My sandbox Support page v2 |
- Currently Asked Questions - »O2k-Open Support alert«
O2k-Open Support resourcesStartup information
Quality control
Advanced topics
Ask an O2k-Expert and the O2k-Open Support agreement
The O2k-Network: An additional resource
|
Funding acknowledgments
|