Difference between revisions of "SUIT-006"
| Line 6: | Line 6: | ||
::: '''[[Categories of SUIT protocols |SUIT-category]]:''' N, S, F, Gp | ::: '''[[Categories of SUIT protocols |SUIT-category]]:''' N, S, F, Gp | ||
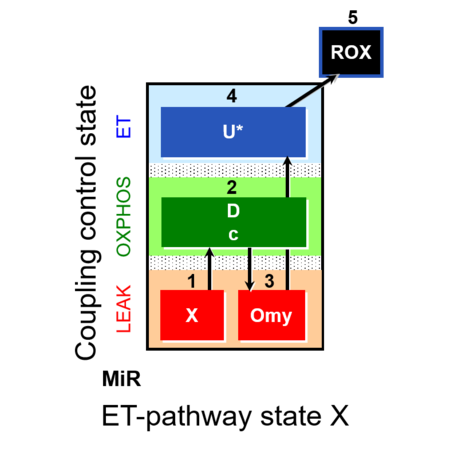
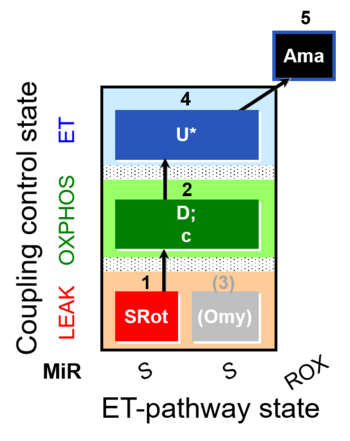
::: '''[[SUIT protocol pattern]]:''' linear [[coupling control protocol]] 1X;2D;3Omy;4U- | ::: '''[[SUIT protocol pattern]]:''' linear [[coupling control protocol]] 1X;2D;3Omy;4U- | ||
The SUIT-006 protocol is a [[coupling control protocol]] for [[mitochondrial preparations]]. Depending on the aims of the researcher, different combinations of substrates can be used to assess coupling control ([[LEAK-respiration|''L'']]-[[Oxidative phosphorylation| ''P'']]-[[ET-capacity| ''E'']] or [[LEAK-respiration|''L'']]-[[Oxidative phosphorylation| ''P'']]-[[LEAK-respiration|''L''(Omy)]]-[[ET-capacity| ''E'']]) at a defined [[electron transfer-pathway state]]. The use of [[oligomycin]] is optional, however, it provides important information when residual and endogenous adenylates are present (which may happen if ATPases are active in the sample). This situation may lead to overestimated [[LEAK]] respiration measured in the abscence of adenylates - [[LEAK state without adenylates|L(n)]]. Therefore, [[oligomycin]] can be used to verify whether this occurs and obtain the | The SUIT-006 protocol is a [[coupling control protocol]] for [[mitochondrial preparations]]. Depending on the aims of the researcher, different combinations of substrates can be used to assess coupling control ([[LEAK-respiration|''L'']]-[[Oxidative phosphorylation| ''P'']]-[[ET-capacity| ''E'']] or [[LEAK-respiration|''L'']]-[[Oxidative phosphorylation| ''P'']]-[[LEAK-respiration|''L''(Omy)]]-[[ET-capacity| ''E'']]) at a defined [[electron transfer-pathway state]]. The use of [[oligomycin]] is optional, however, it provides important information when residual and endogenous adenylates are present (which may happen if ATPases are active in the sample). This situation may lead to overestimated [[LEAK]] respiration measured in the abscence of adenylates - [[LEAK state without adenylates|L(n)]]. Therefore, [[oligomycin]] can be used to verify whether this occurs and obtain the [[LEAK]] state appropriately. | ||
Revision as of 19:17, 7 March 2019
Description
Abbreviation: N, S, F, Gp
Reference: A: Coupling control protocol, mtprep ![]() »Versions
»Versions
- SUIT-category: N, S, F, Gp
- SUIT protocol pattern: linear coupling control protocol 1X;2D;3Omy;4U-
The SUIT-006 protocol is a coupling control protocol for mitochondrial preparations. Depending on the aims of the researcher, different combinations of substrates can be used to assess coupling control (L- P- E or L- P-L(Omy)- E) at a defined electron transfer-pathway state. The use of oligomycin is optional, however, it provides important information when residual and endogenous adenylates are present (which may happen if ATPases are active in the sample). This situation may lead to overestimated LEAK respiration measured in the abscence of adenylates - L(n). Therefore, oligomycin can be used to verify whether this occurs and obtain the LEAK state appropriately.
Communicated by Cardoso LH, Doerrier C, Gnaiger E, Iglesias-Gonzalez J and Komlodi T (last update 2019-03-07)
Specific SUIT protocols
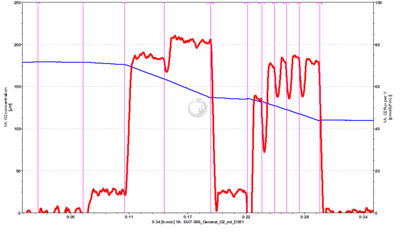
- SUIT-006 General O2 mt D051 General coupling protocol for isolated mitochondria and tissue homogenates. This general protocol is designed to study the coupling control state of mitochondrial preparations (isolated mitochondria and tissue homogenates). Different substrate/inhibitor combinations (X) can be used. The use of oligomycin is optional depending on if we are interested in measuring LEAK state.
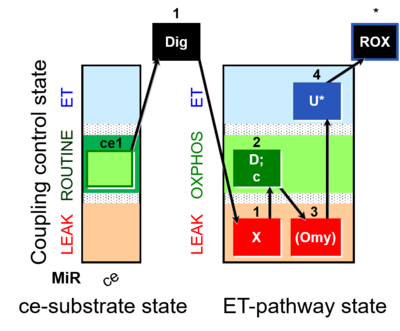
- SUIT-006 General O2 pce D053: General coupling protocol for permeabilized cells. This general protocol is designed to study the coupling control state of permeabilized cells. Different substrate/inhibitor combinations (X) can be used. The use of oligomycin is optional depending on if we are interested in measuring LEAK state.
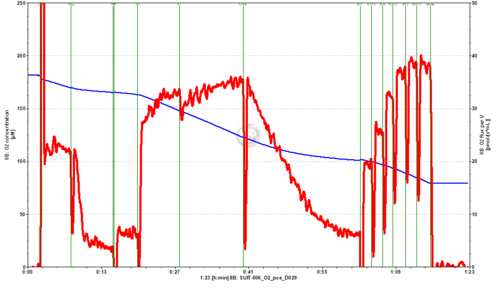
- SUIT-006 O2 pce D029 for N-pathway with permeabilized cells. Omy may be added in order to not overestimate LEAK respiration, that might be overestimated in 1PM if there are endogenous adenylates.
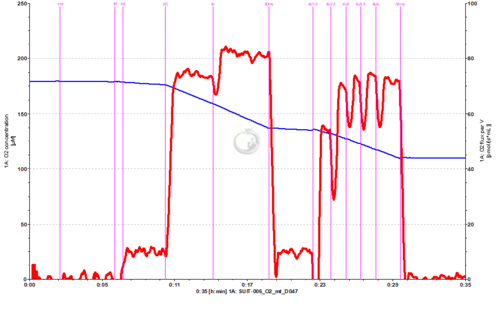
- SUIT-006 O2 mt D047 for N-pathway with isolated mitochondria or tissue homogenate. Omy may be added in order to not overestimate LEAK respiration, that might be overestimated in 1PM if there are endogenous adenylates.
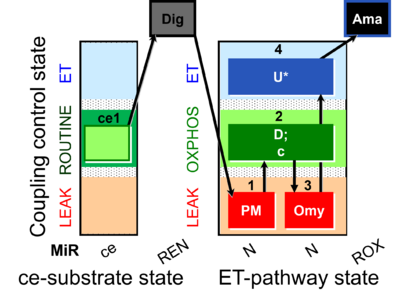
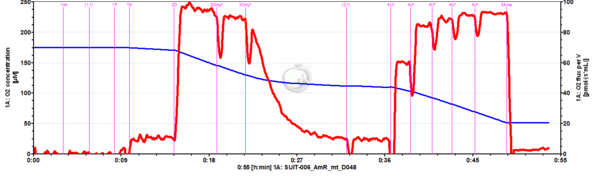
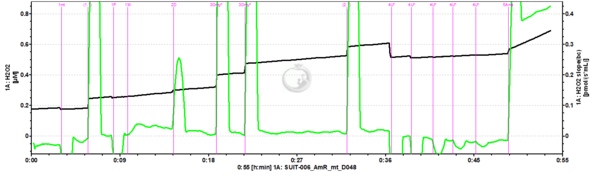
- SUIT-006 AmR mt D048 to study simultaneously the O2 flux and H2O2 flux on isolated mitochondria and tissue homogenate, in particular to study the influence of mt-membrane potential on NADH-linked pathway initiated mt-H2O2 production.
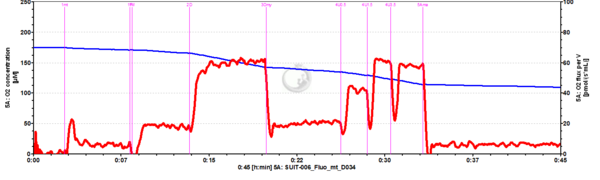
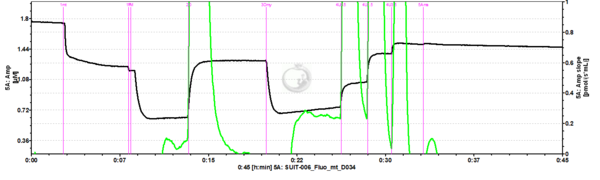
- SUIT-006 Fluo mt D034 to study simultaneously the O2 flux and mitochondrial membrane potential on isolated mitochondria and tissue homogenate.
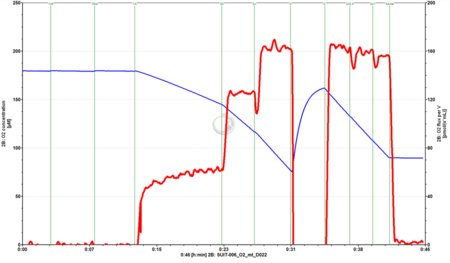
- SUIT-006 O2 mt D022 for S-pathway with isolated mitochondria or tissue homogenate.
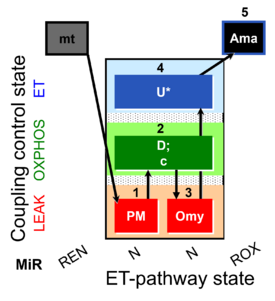
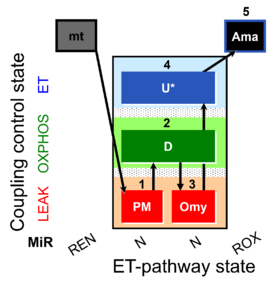
Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 1X | XL(n) | 1X
| ||
| 2D | XP | 1X;2D
| ||
| 2c | XcP | 1X;2D;2c
| ||
| 3Omy | XLOmy | 1X;2D;2c;3Omy
| ||
| 4U | XE | 1X;2D;2c;3Omy;4U
| ||
| 5Ama | ROX | 1X;2D;2c;3Omy;4U;5Ama
|
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary
Strengths and limitations
- This protocol is indicated when the aim is to analyse coupling control in mtprep (see MitoPedia: Sample preparations) at a defined electron transfer-pathway state.
- + Different combinations of substrates and inhibitors (X) can be used depending on the specific aims, e.g.: PGM, PM, GM, MnaPM or other combinations for N-pathway control state; S or RotS for S-pathway control state.
- + The combinations of substrates for one ET-pathway state with inhibitors for other pathways provide the best coupling control protocols for one specific ET-pathway state, e.g.: PM in the presence of Mna for N-pathway control state, succinate in the presence of rotenone for S-pathway control state. In the specific case of S-pathway control state, rotenone should be added to avoid reverse electron transfer (RET).
- + Reasonable duration of the experiment.
- + This protocol can be extended with the Complex IV module.
- - This protocol does not include internal ET-pathway control steps.
- - Careful washing is required after the experiment to avoid carry-over of uncoupler and inhibitors, if applied (e.g., rotenone&succinate, SRot-pathway control state).
- - LEAK respiration in the absence of Omy may be overestimated due to ATP turnover stimulated by endogenous adenylates. This can be evaluated by SUIT 1X;2D;3Omy;4U;-. In mouse brain homogenate with succinate&rotenone, L(n) and L(Omy) are identical (Krumschnabel et al 2014).
- - CIV activity and Cytochrome c test cannot be performed together with the fluorescence (SUIT-006 AmR mt D048 and SUIT-006 AmR mt D034).
Compare SUIT protocols
- SUIT-001: Extended protocol (Reference protocol 1) with SUIT-006 as the initial coupling control module (with PM as NADH-linked substrates).
- SUIT-004: Shortened version of SUIT-001, also with SUIT-006 as the initial coupling control module (with PM as NADH-linked substrates).
- SUIT-003: Coupling control protocol for non-permeabilized cells (ce), counterpart to SUIT-006 for mt-preparations (mtprep).
References
| Year | Reference | Organism | Tissue;cell | |
|---|---|---|---|---|
| MiPNet26.10 MgG data analysis | 2021-09-16 | Measurement of mitochondrial ATP production using Magnesium Green: DL-Protocols and data analysis with DatLab 7.4 | ||
| Komlodi 2021 BEC AmR-O2 | 2021 | Komlódi T, Sobotka O, Gnaiger E (2021) Facts and artefacts on the oxygen dependence of hydrogen peroxide production using Amplex UltraRed. Bioenerg Commun 2021.4. https://doi.org/10.26124/bec:2021-0004 | Saccharomyces cerevisiae | Other cell lines |
| Cardoso 2021 BEC MgG | 2021 | Cardoso LHD, Doerrier C, Gnaiger E (2021) Magnesium Green for fluorometric measurement of ATP production does not interfere with mitochondrial respiration. Bioenerg Commun 2021.1. https://doi.org/10.26124/bec:2021-0001 | Mouse | Heart |
| Krumschnabel 2014 Methods Enzymol | 2014 | Krumschnabel G, Eigentler A, Fasching M, Gnaiger E (2014) Use of safranin for the assessment of mitochondrial membrane potential by high-resolution respirometry and fluorometry. Methods Enzymol 542:163-81. https://doi.org/10.1016/B978-0-12-416618-9.00009-1 | Mouse | Nervous system |
MitoPedia concepts:
MiP concept,
SUIT protocol,
Recommended
MitoPedia methods:
Respirometry,
Fluorometry