Difference between revisions of "Template:SUIT-007 pce"
From Bioblast
Beno Marija (talk | contribs) |
|||
| (5 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
[[Image:SUIT-MitoFit.png|right|190px|link=http://www.bioblast.at/index.php/MitoPedia:_SUIT |MitoPedia: SUIT]] | [[Image:SUIT-MitoFit.png|right|190px|link=http://www.bioblast.at/index.php/MitoPedia:_SUIT |MitoPedia: SUIT]] | ||
== Steps and respiratory states == | == Steps and respiratory states == | ||
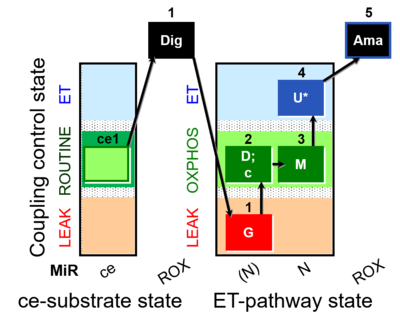
[[File:Ce1;1Dig;1G;2D;2c;3M;4U | [[File:Ce1;1Dig;1G;2D;2c;3M;4U;5Ama.png|400px]] | ||
{{Template:SUIT cells}} | {{Template:SUIT cells}} | ||
| Line 10: | Line 10: | ||
|- | |- | ||
| 1G | | 1G | ||
| [[Glutamate|G]]<sub>''L''</sub> | | [[Glutamate|G]]<sub>''[[LEAK respiration|L]]''</sub> | ||
| ([[N]]) | | ([[N]]) | ||
| CI | | CI | ||
| 1G | | 1G | ||
*N<sub>''L''</sub> if [[glutamate dehydrogenase]] is present in the sample studied ([[Glutamate anaplerotic pathway control state]]). | *N<sub>''L''(n)</sub> if [[glutamate dehydrogenase]] is present in the sample studied ([[Glutamate-anaplerotic pathway control state]]). | ||
*{{Template:SUIT L(n)}} | *{{Template:SUIT L(n)}} | ||
|- | |- | ||
| 2D | | 2D | ||
| [[Glutamate|G]]<sub>''P''</sub> | | [[Glutamate|G]]<sub>''[[OXPHOS|P]]''</sub> | ||
| ([[N]]) | | ([[N]]) | ||
| CI | | CI | ||
| 1G;2D | | 1G;2D | ||
*N<sub>''P''</sub> if [[glutamate dehydrogenase]] is present in the sample studied ([[Glutamate anaplerotic pathway control state]]). | *N<sub>''[[OXPHOS|P]]''</sub> if [[glutamate dehydrogenase]] is present in the sample studied ([[Glutamate-anaplerotic pathway control state]]). | ||
*{{Template:SUIT OXPHOS}} | *{{Template:SUIT OXPHOS}} | ||
|- | |- | ||
| 2c | | 2c | ||
| [[Glutamate|G]]c<sub>''P''</sub> | | [[Glutamate|G]]c<sub>''[[OXPHOS|P]]''</sub> | ||
| ([[N]]) | | ([[N]]) | ||
| CI | | CI | ||
| Line 40: | Line 40: | ||
|- | |- | ||
| 3M | | 3M | ||
| [[GM]]<sub>''P''</sub> | | [[GM]]<sub>''[[OXPHOS|P]]''</sub> | ||
| [[N]] | | [[N]] | ||
| CI | | CI | ||
| Line 50: | Line 50: | ||
|- | |- | ||
| 4U | | 4U | ||
| [[GM]]<sub>''E''</sub> | | [[GM]]<sub>''[[E]]''</sub> | ||
| [[N]] | | [[N]] | ||
| CI | | CI | ||
| Line 63: | Line 63: | ||
| | | | ||
| | | | ||
| | | 1G;2D;2c;3M;4U;5Ama | ||
*{{Template:SUIT Ama}} | *{{Template:SUIT Ama}} | ||
|} | |} | ||
{{Keywords: SUIT protocols}} | |||
{{ | |||
Latest revision as of 13:10, 25 November 2020
Steps and respiratory states
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| ce1 | ROUTINE | ce1
| ||
| 1Dig | REN | ce1;1Dig
|
| Step | State | Pathway | Q-junction | Comment - Events (E) and Marks (M) |
|---|---|---|---|---|
| 1G | GL | (N) | CI | 1G
|
| 2D | GP | (N) | CI | 1G;2D
|
| 2c | GcP | (N) | CI | 1G;2D;2c
|
| 3M | GMP | N | CI | 1G;2D;2c;3M
|
| 4U | GME | N | CI | 1G;2D;2c;3M;4U
|
| 5Ama | ROX | 1G;2D;2c;3M;4U;5Ama
|
- Bioblast links: SUIT protocols - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Coupling control
- Pathway control
- Main fuel substrates
- » Glutamate, G
- » Glycerophosphate, Gp
- » Malate, M
- » Octanoylcarnitine, Oct
- » Pyruvate, P
- » Succinate, S
- Main fuel substrates
- Glossary