From Bioblast
Revision as of 19:18, 17 September 2016 by Gnaiger Erich (talk | contribs) (moved MitoPedia: SUIT all to MitoPedia: SUIT A)
The MitoPedia terminology is developed continuously in the spirit of Gentle Science.
- MitoPedia: SUIT is a component of OROBOROS support.
- In the Library of SUIT protocols
- » MitoPedia: SUIT A - these are SUIT protocols used for cohort studies.
- » MitoPedia: SUIT B - these are published SUIT protocols considered less suitable for cohort studies.
- » MitoPedia: SUIT C - these are SUIT protocols used for exploratory research.
- » MitoPedia: SUIT - you are here: this is a list of all SUIT protocols in the library of SUIT protocols.
- See also:
- » O2k-Protocols: O2k-Demo experiments
- In the Library of SUIT protocols
Library of SUIT protocols: all
| Short name | Systematic name | Coupling/pathway control diagram | Reference |
|---|---|---|---|
| 1OctM;2D;3PG;4S;5U;6Rot- | FNS(Oct,PGM) | 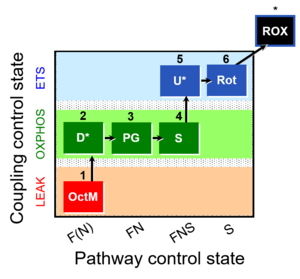 | C MiPNet18.13 IOC84 Alaska |
| 1PGM;2D;3S;4Rot;5U- | NS(PGM) | 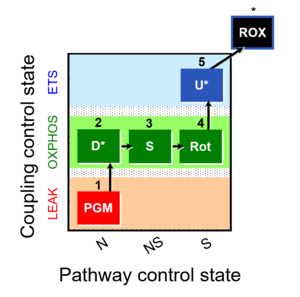 | C |
| 1PGM;2D;3U;4S;5Rot- | NS(PGM) | 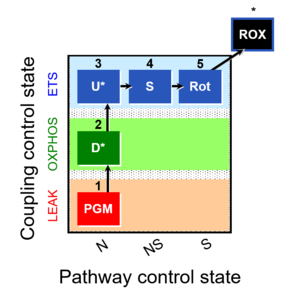 | C MiPNet18.13 IOC84 Alaska |
| 1PM;2D;3G;4U;5S;6Rot- | NS(PGM) | 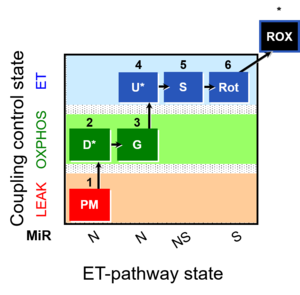 | B Lemieux 2011 Int J Biochem Cell Biol |
| SUIT-001 | RP1 | 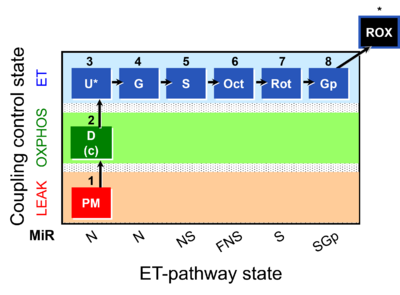 | Doerrier 2018 Methods Mol Biol |
| SUIT-001 O2 ce-pce D003 | RP1 ce-pce | 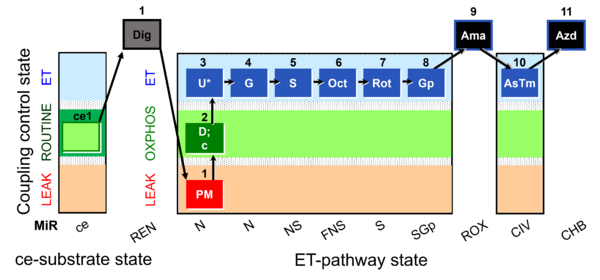 | A - SUIT-001 - SUIT reference protocol Reference protocol RP1 for living cells (ce, ROUTINE respiration) and cells permeabilized in the respirometric chamber (pce) |
| SUIT-001 O2 ce-pce D004 | RP1 ce-pce blood | 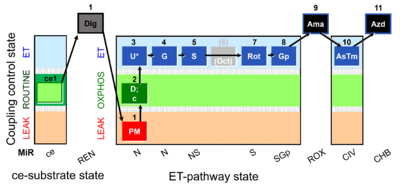 | A - SUIT-001 - SUIT reference protocol Reference protocol RP1 for living PBMC and PLT cells (ce, ROUTINE respiration) and cells that are permeabilized in the chamber (pce) |
| SUIT-001 O2 mt D001 | RP1 mt | 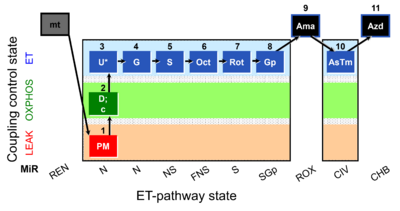 | SUIT-001 - SUIT reference protocol Reference protocol RP1 for isolated mitochondria, tissue homogenate and permeabilized cells |
| SUIT-001 O2 pfi D002 | RP1 pfi | 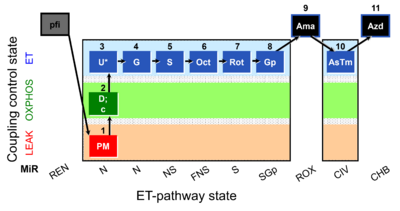 | SUIT-001 - SUIT reference protocol Reference protocol RP1 for permeabilized fibers |
| SUIT-002 | RP2 | 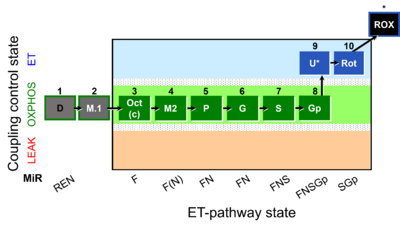 | A: Doerrier 2018 Methods Mol Biol |
| SUIT-002 O2 ce-pce D007 | RP2 ce-pce | 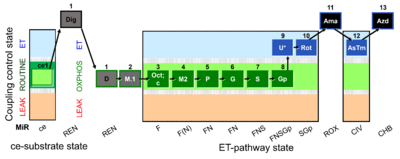 | A - SUIT-002 - SUIT reference protocol Reference protocol RP2 for living cells (ce, ROUTINE respiration) and cells that are permeabilized in the chamber (pce) |
| SUIT-002 O2 ce-pce D007a | RP2 ce-pce blood |  | A - SUIT-002 - SUIT reference protocol Reference protocol RP2 for for living PBMC and PLT cells (ce, ROUTINE respiration) and cells that are permeabilized in the chamber (pce) |
| SUIT-002 O2 mt D005 | RP2 mt | 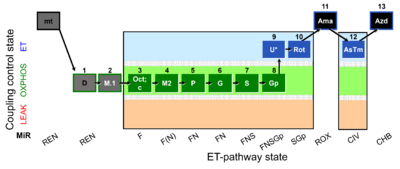 | A - SUIT-002 - SUIT reference protocol Reference protocol RP2 for isolated mitochondria, tissue homogenate and permeabilized cells |
| SUIT-002 O2 pfi D006 | RP2 pfi | 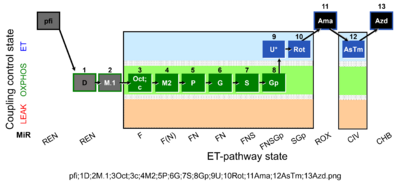 | A -SUIT-002 - SUIT reference protocol Reference protocol RP2 for permeabilized fibers (pfi) |
| SUIT-003 | CCP-ce | 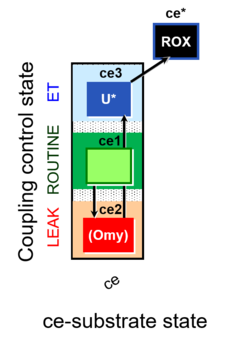 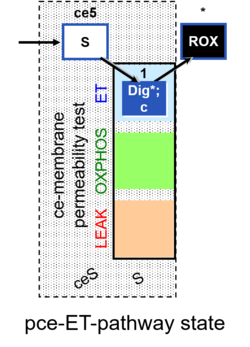 | A: Coupling-control protocol, living cells »MiPNet08.09 CellRespiration Bioblast pdf |
| SUIT-003 AmR ce D017 | CCP-ce S permeability test | 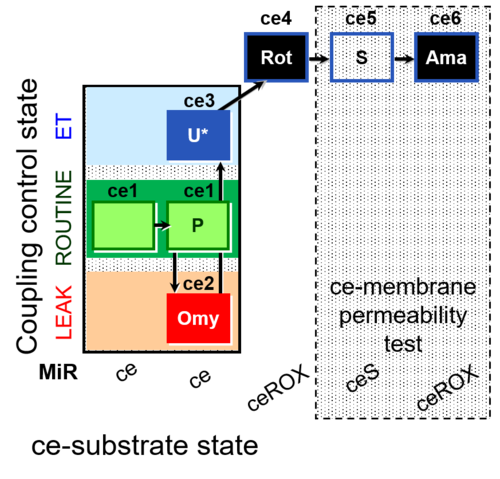 | B: CCP(P) and cell membrane permeability test with succinate - SUIT-003 |
| SUIT-003 AmR ce D058 | AmR effect on ce | 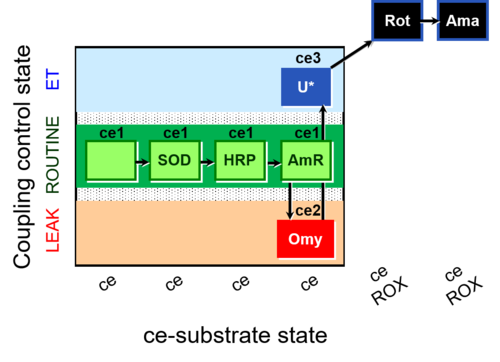 | B: Coupling control in intact cells with oligomycin to study the inhibitory effect of the AmR assay components on the respiration ; ce: non-permeabilized cells -SUIT-003 |
| SUIT-003 AmR ce D059 | AmR effect on ce - control | 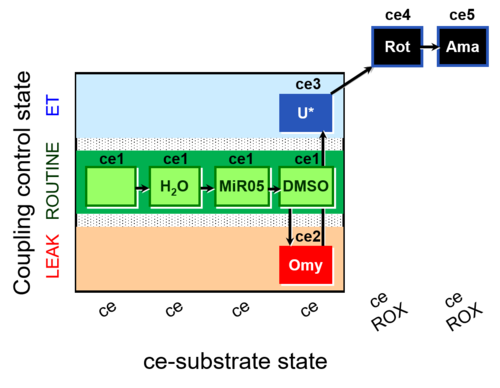 | B: Experimental control for SUIT-003 AmR ce D058 |
| SUIT-003 Ce1;ce1P;ce3U;ce4Glc;ce5M;ce6Rot;ce7S;1Dig;1c;2Ama;3AsTm;4Azd | cePMGlc,S | 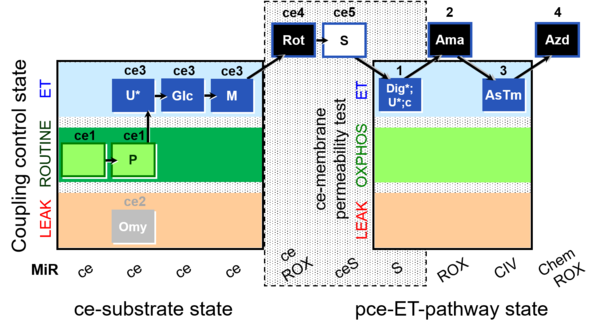 | C SUIT-003 Coupling-control protocol and respirometric cell viability test (CCV protocol) without oligomycin. |
| SUIT-003 Ce1;ce1SD;ce2Omy;ce3U- | FNS(Oct,PGM) | 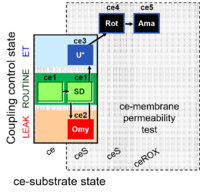 | B SUIT-003 Stadlmann 2006 Cell Biochem Biophys |
| SUIT-003 Ce1;ce1SD;ce3U;ce4Rot;ce5Ama | ceS | 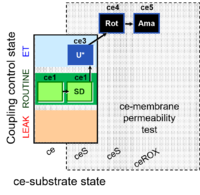 | B SUIT-003 CCP(S) |
| SUIT-003 Ce1;ce2SD;ce3Omy;ce4U- | FNS(Oct,PGM) | 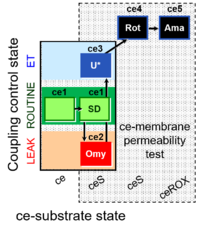 | B SUIT-003 Stadlmann 2006 Cell Biochem Biophys |
| SUIT-003 Ce1;ce2SD;ce3U;ce4Rot;ce5Ama | ceS | 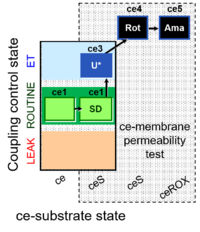 | B Steinlechner-Maran 1997 Transplantation |
| SUIT-003 Ce1;ce2U;ce3Rot;ce4S;ce5Ama | ceS | 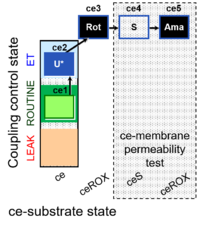 | B SUIT-003 CCP(S) |
| SUIT-003 Ce1;ce3U;ce4Rot;ce5S;ce6Ama | ceS | 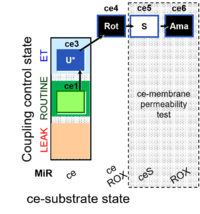 | B SUIT-003 CCP(S) |
| SUIT-003 O2 ce D009 | CCP-ce short | 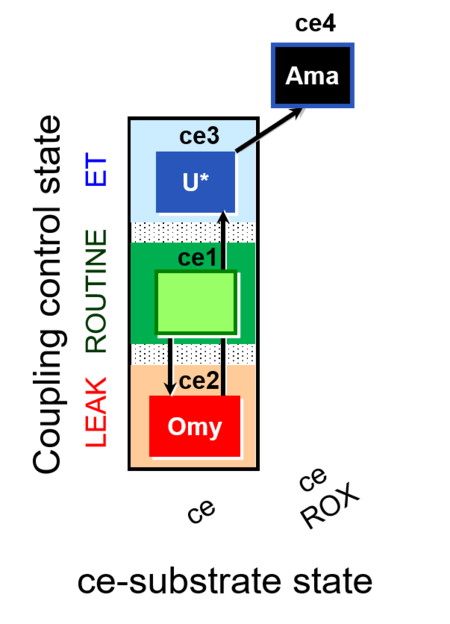 | A: - SUIT-003 - Coupling-control protocol (CCP) in living cells (ce) with oligomycin (LEAK state) |
| SUIT-003 O2 ce D012 | CCP-ce(P) | 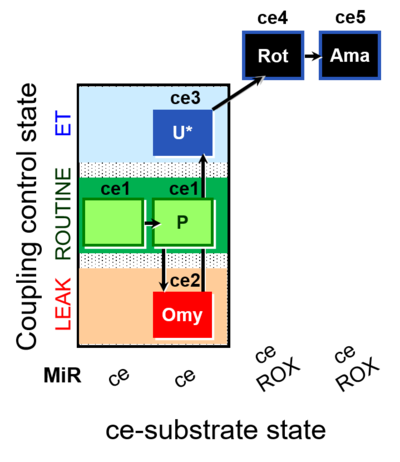 | B - SUIT-003 |
| SUIT-003 O2 ce D028 | CCP-ce S permeability test | 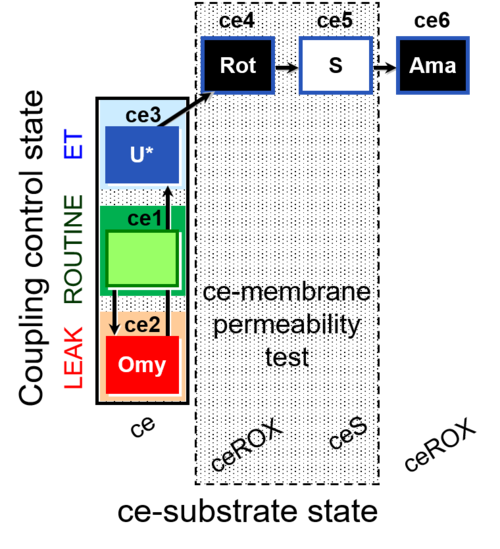 | B SUIT-003 Coupling-control protocol with succinate for permeability test |
| SUIT-003 O2 ce D037 | CCP-ce Crabtree_R | 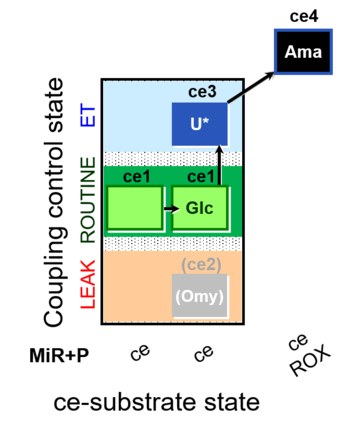 | A SUIT-003 - Coupling-control protocol and Crabtree effect |
| SUIT-003 O2 ce D038 | CCP-ce Crabtree_E | 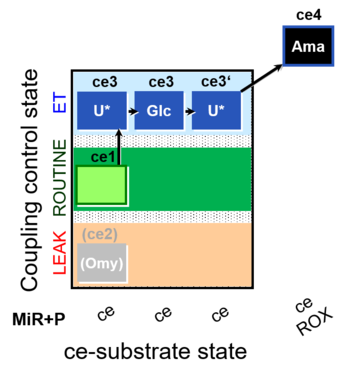 | A Coupling-control protocol and Crabtree effect test -SUIT-003 complex cell coupling-control protocol |
| SUIT-003 O2 ce D039 | CCP-ce microalgae | 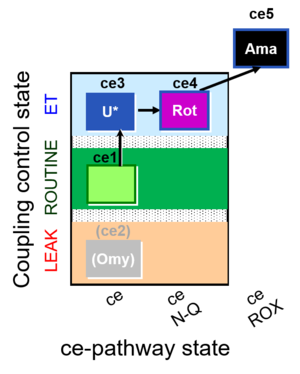 | A: Coupling control in intact cells without oligomycin; ce: non-permeabilized cells -SUIT-003 |
| SUIT-003 O2 ce D050 | CCP-ce Snv | 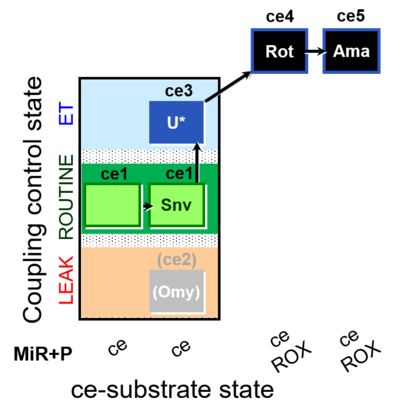 | A: - Coupling-control protocol for living cells with plasma membrane-permeable succinate (MitoKit-CII/Succinate-nv) - SUIT-003 - Plasma membrane-permeable succinate (NV118) in ROUTINE respiration. Coupling-control protocol (CCP) in living cells (ce) |
| SUIT-003 O2 ce D060 | CCP-ce Snv,Mnanv | 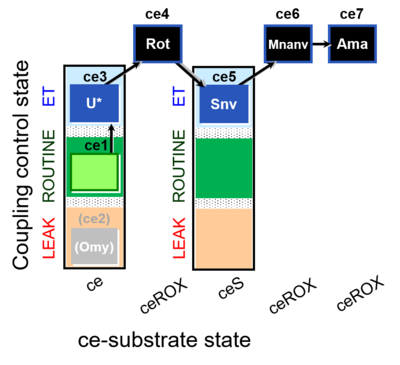 | B - coupling-control protocol for living cells with cell permeable compounds MitoKit-CII/Succinate-nv and MitoKit-CII/Malonate-nv -SUIT-003 - Coupling-control protocol (CCP) with plasma membrane-permeable succinate (MitoKit-CII/Succinate-nv) and plasma membrane-permeable malonate (MitoKit-CII/Malonate-nv) in living cells (ce). |
| SUIT-003 O2 ce D061 | CCP-ce Snv,Mnanv - control | 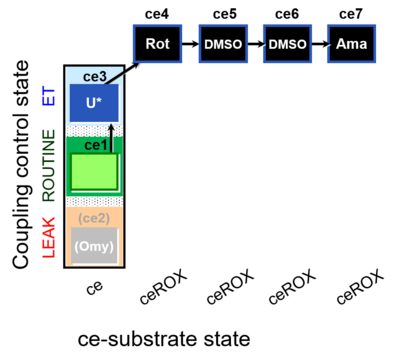 | B Carrier control for the coupling-control protocol for living cells with cell permeable compounds MitoKit-CII/Succinate-nv and MitoKit-CII/Malonate-nv - SUIT-003 O2 ce D060 |
| SUIT-003 O2 ce D062 | CCP-ce Snv - control | 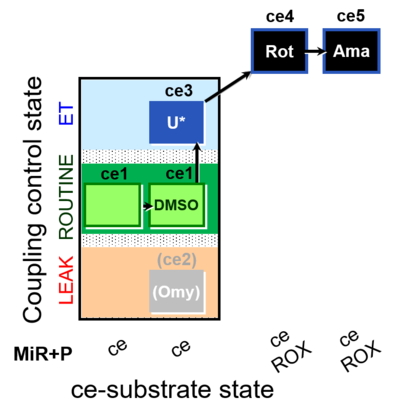 | A: Carrier control for the Coupling-control protocol for living cells with plasma membrane-permeable succinate (MitoKit-CII/Succinate-nv) - SUIT-003 O2 ce D050 |
| SUIT-003 O2 ce-pce D013 | CCVP-Glc,M | 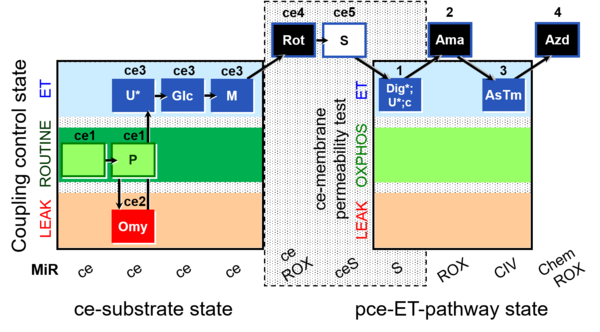 | C - SUIT-003 |
| SUIT-003 O2 ce-pce D018 | CCVP-Glc | 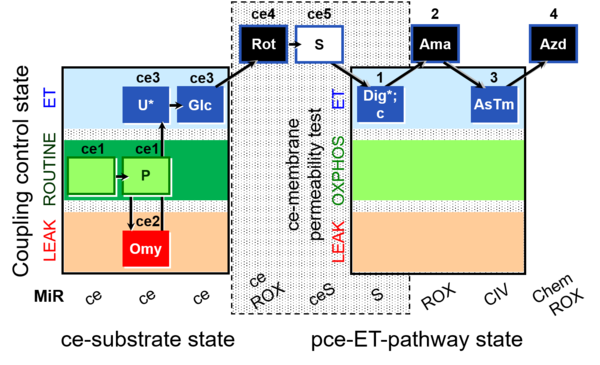 | C SUIT-003 Second version of the Coupling-control protocol and respirometric cell viability test (CCV protocol) |
| SUIT-003 O2 ce-pce D020 | CCVP | 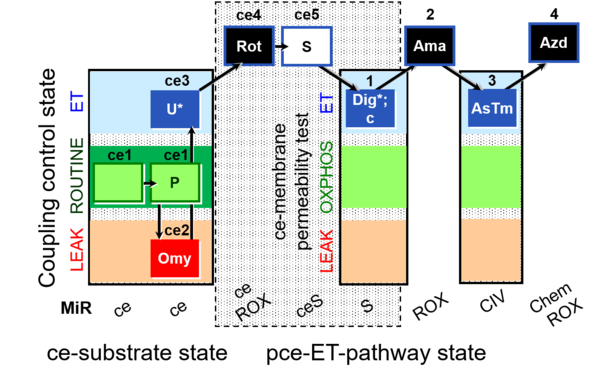 | A SUIT-003 - Coupling-control protocol and respirometric cell viability test; ce: living cells, pce: permeabilized cells |
| SUIT-003 pH ce D067 | CCP-Crabtree with glycolysis inhibition | 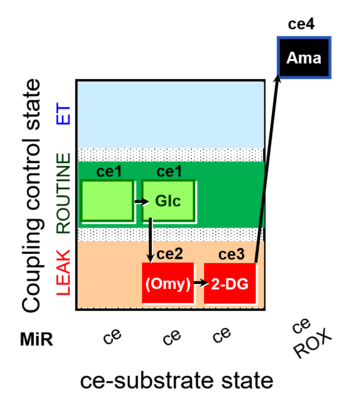 | A Coupling-control protocol and Crabtree effect with glycolysis inhibition- SUIT-003 |
| SUIT-004 | RP1-short | 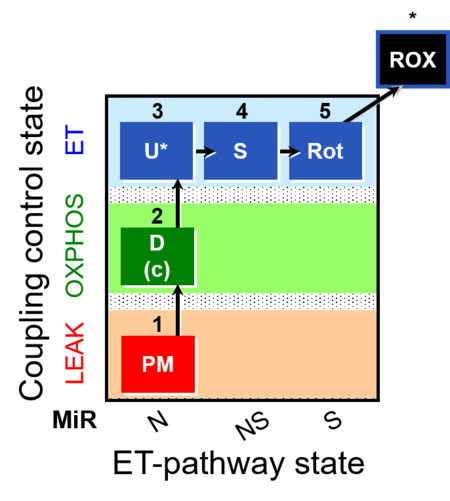 | |
| SUIT-004 O2 pfi D010 | RP1-short pfi | 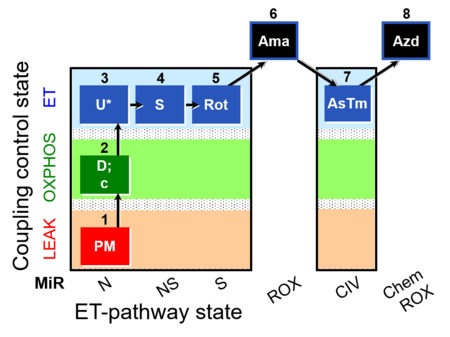 | SUIT-004 - Short protocol linked to SUIT-001 O2 pfi D002 SUIT RP1 (human skeletal muscle) |
| SUIT-005 | RP2-short | 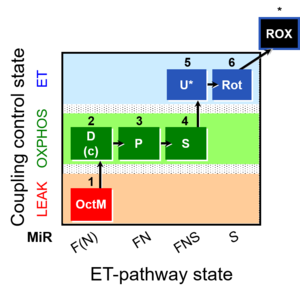 | A: when malate-anaplerotic activity is zero |
| SUIT-005 O2 pfi D011 | RP2-short pfi | 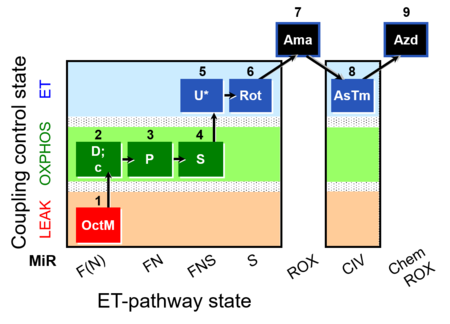 | A: - SUIT-005 short protocol linked to SUIT-002 O2 pfi D006 - SUIT RP2 (human skeletal muscle) |
| SUIT-006 | CCP-mtprep | 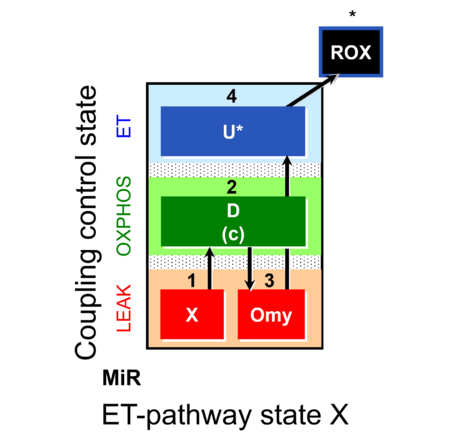 | A: Coupling-control protocol, mtprep |
| SUIT-006 AmR mt D048 | CCP mt PM - AmR | 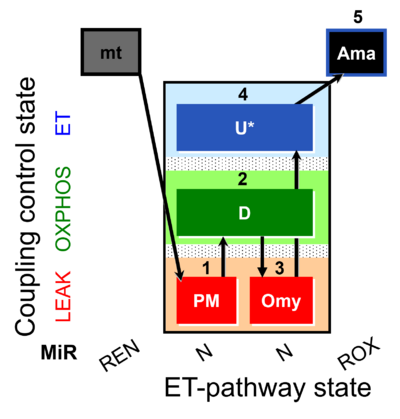 | A: short protocol for simultaneous determination of O2 flux and H2O2 in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells)- SUIT-006 |
| SUIT-006 Fluo mt D034 | CCP mt PM - Fluo | 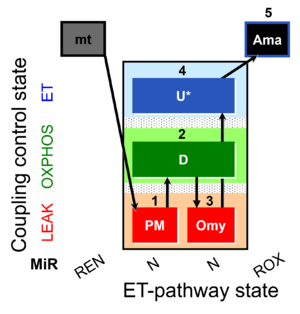 | A: short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) -SUIT-006 |
| SUIT-006 MgG ce-pce D085 | CCP MgG ce-pce | 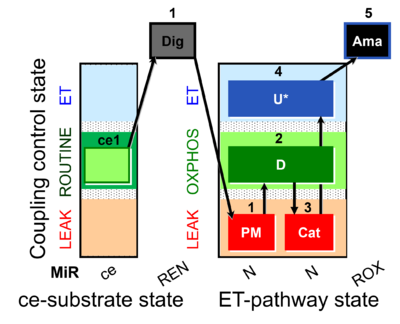 | A: short coupling-control protocol for simultaneous determination of O2 flux and mitochondrial ATP production in permeabilized cells -SUIT-006 |
| SUIT-006 MgG mt D055 | CCP MgG mt | 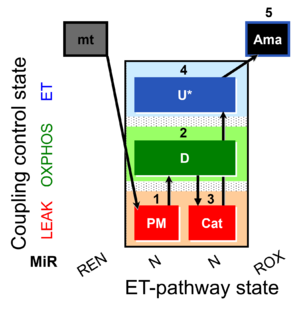 | A: short coupling-control protocol for simultaneous determination of O2 flux and mitochondrial ATP production in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) -SUIT-006 |
| SUIT-006 O2 ce-pce D029 | CCP ce-pce PM | 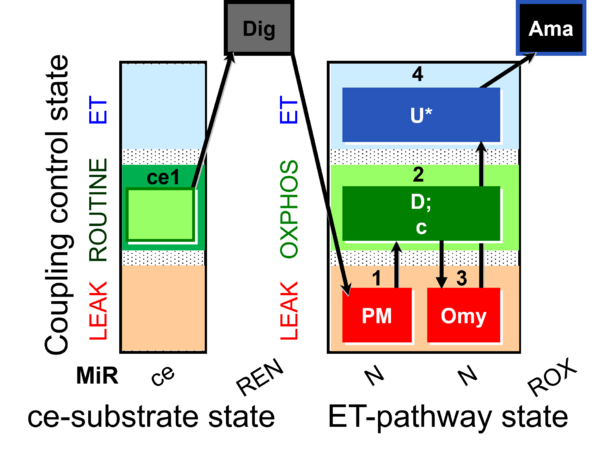 | A to analyse the coupling control on permeabilized cells in N(PM) pathway control state- SUIT-006 |
| SUIT-006 O2 mt D022 | CCP mt S(Rot) | 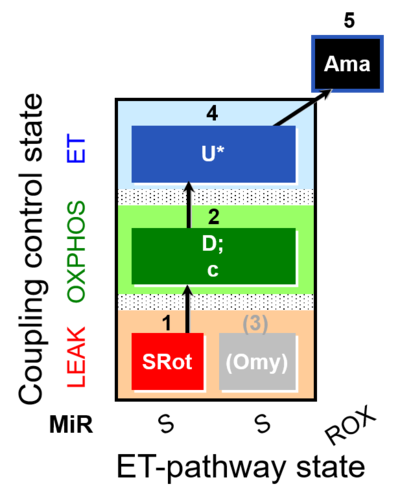 | A - SUIT-006 - Protocol for coupling control assessment in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) |
| SUIT-006 O2 mt D047 | CCP mt PM | 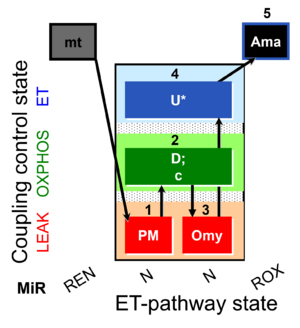 | A SUIT-006 - is a coupling-control protocol in the N-pathway for isolated mitochondria, tissue homogenate and cells permeabilized before addition to the O2k chamber. |
| SUIT-006 Q ce-pce D073 | CCP ce-pce S(Rot) | 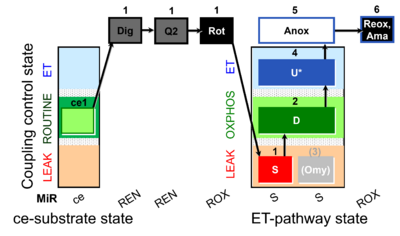 | A Coupling control protocol to analyze mitochondrial O2 consumption and the redox state of the Q-pool in S(Rot)-pathway control state in permeabilized cells- SUIT-006 |
| SUIT-006 Q mt D071 | CCP mt S(Rot) | 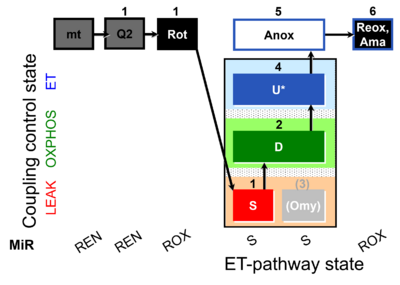 | A Coupling-control protocol to analyze mitochondrial O2 consumption and the redox state of the Q-pool in S(Rot)-pathway control state in isolated mitochondria- SUIT-006 |
| SUIT-007 | Glutamate anaplerosis | 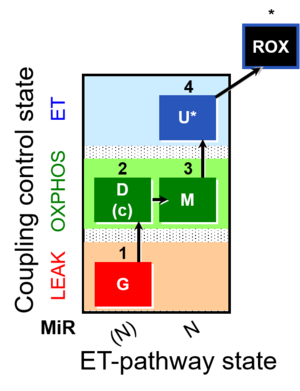 | A: Glutamate anaplerotic pathway |
| SUIT-007 O2 ce-pce D030 | Glutamate anaplerotic pathway | 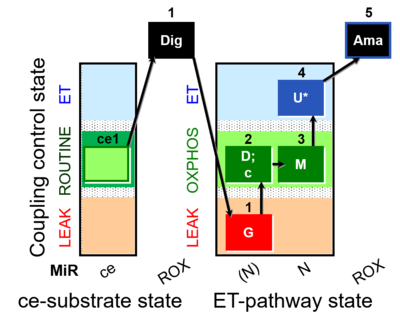 | A pce: permeabilized cells - SUIT-007 |
| SUIT-008 | PM+G+S_OXPHOS+Rot_ET | 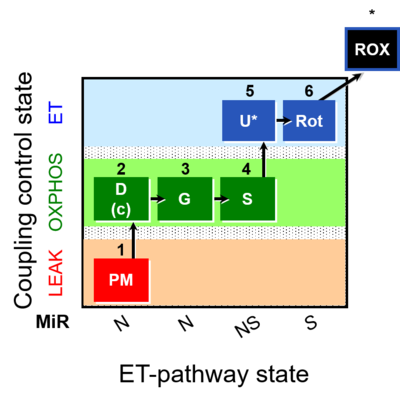 | A: Additivity between the N- and S-pathway in the Q-junction |
| SUIT-008 O2 ce-pce D025 | Q-junction ce-pce | 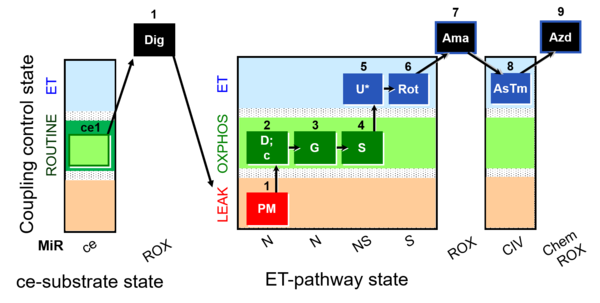 | A: for cells-permeabilized cells Lemieux 2017 Sci Rep - SUIT-008 |
| SUIT-008 O2 mt D026 | Q-junction mtprep | 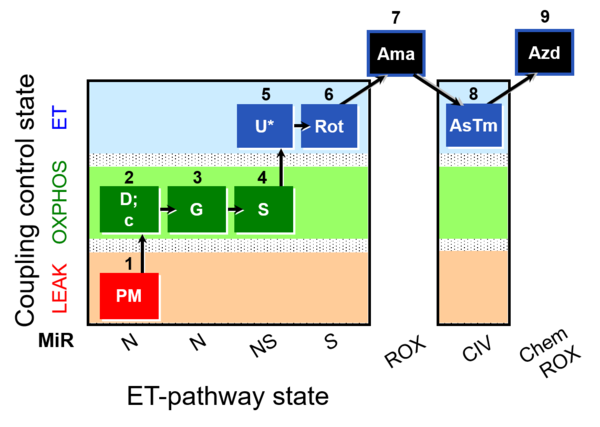 | A: SUIT protocol for mt; mt: isolated mitochondria and tissue homogenate - SUIT-008 |
| SUIT-008 O2 pce D25 | NS(PGM) | 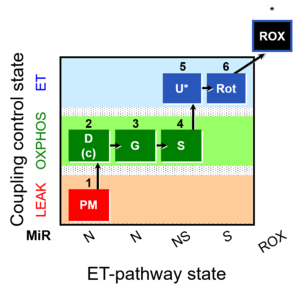 | B: SUIT 8 for permeabilized cells Lemieux 2017 Sci Rep |
| SUIT-008 O2 pfi D014 | 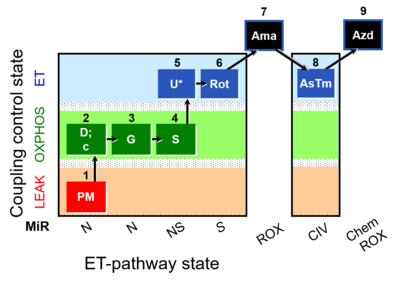 | A - Lemieux 2017 Sci Rep - SUIT-008- O2 pfi D014 protocol was used a a reference protocol in MitoEAGLE project (WG2) to evaluate the researcher skills preparing permeabilized fibers | |
| SUIT-009 AmR ce-pce D019 | H2O2 RET ce-pce S_L | 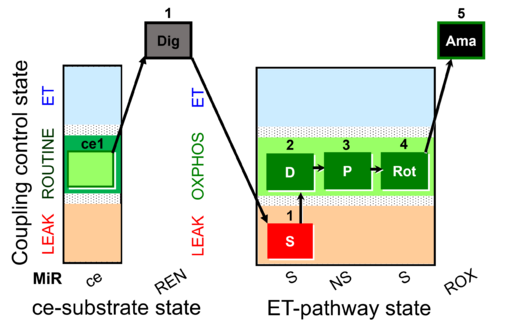 | B: short protocol for H2O2 (AmR) in ce: non-permeabilized cells, pce: permeabilized cells -SUIT-009 |
| SUIT-009 AmR mt D021 | H2O2 mtprep | 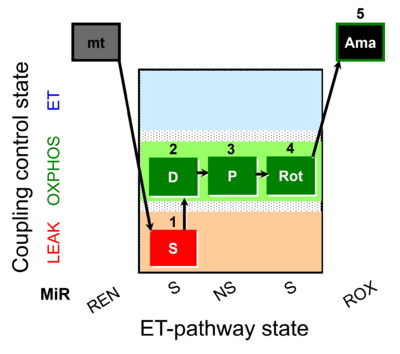 | B: short protocol for H2O2 (AmR) in mitochondrial preparations; mt: isolated mitochondria, tissue homogenate and permeabilized cells - SUIT-009short protocol for H2O2 (AmR) for isolated mitochondria, tissue homogenate and permeabilized cells |
| SUIT-009 O2 ce-pce D016 | 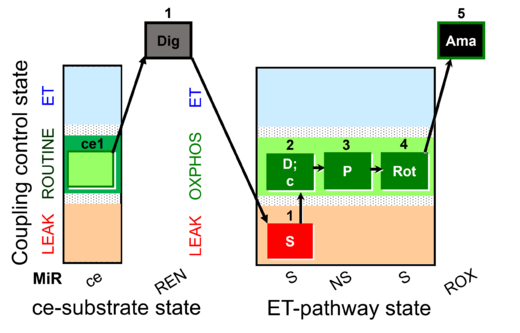 | B - SUIT-009 - short protocol for mt-respiration in ce: non-permeabilized cells, pce: permeabilized cells. | |
| SUIT-009 O2 mt D015 | 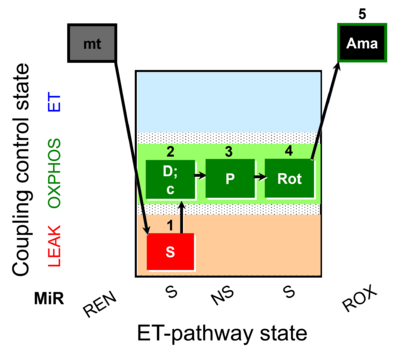 | B - SUIT-009- short protocol for respiration in mitochondrial preparations; mt: isolated mitochondria, tissue homogenate and permeabilized cells | |
| SUIT-010 | Digitonin test | 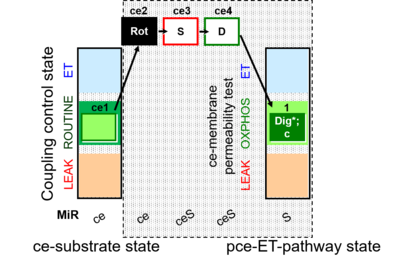 | A: Optimization of digitonin concentration for pce |
| SUIT-010 O2 ce-pce D008 | Dig titration-pce |  | A -SUIT-010 - Optimum digitonin concentration for pce |
| SUIT-011 | GM+S_OXPHOS+Rot_ET | 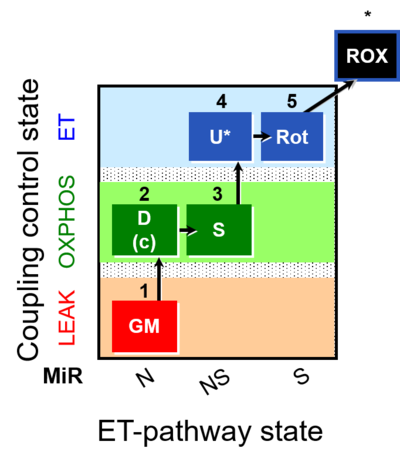 | A: Maximum mitochondrial respiratory capacity (OXPHOS with NS substrates) and coupling/pathway control |
| SUIT-011 O2 pfi D024 | NS physiological maximum capapcity in fibres | 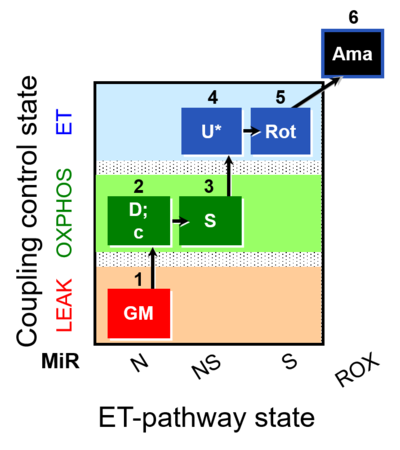 | A pfi: permeabilized fibers - SUIT-011 |
| SUIT-012 | PM+G_OXPHOS | 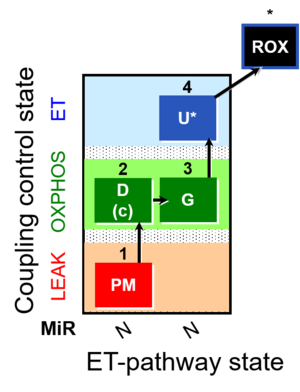 | A: Coupling control (L- P- E) with NADH-linked substrates (PM and PGM) |
| SUIT-012 O2 ce-pce D052 | N(PGM) | 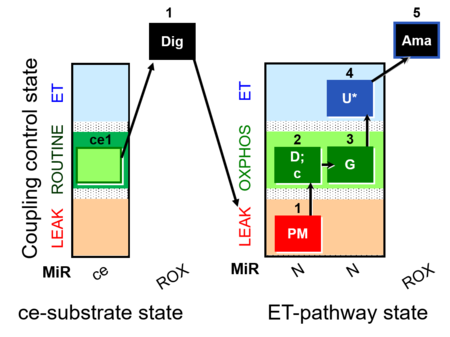 | A Linear coupling control with NADH-linked substrates for pce - SUIT-012 |
| SUIT-012 O2 mt D027 | N CCP mtprep | 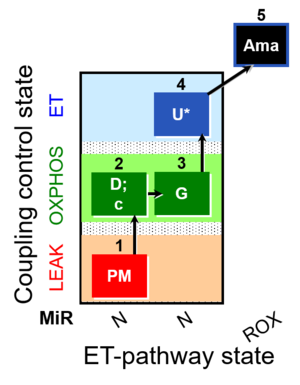 | A Linear coupling control with NADH-linked substrates for isolated mitochondria, tissue homogenate and permeabilized cells- SUIT-012 |
| SUIT-013 | ce | 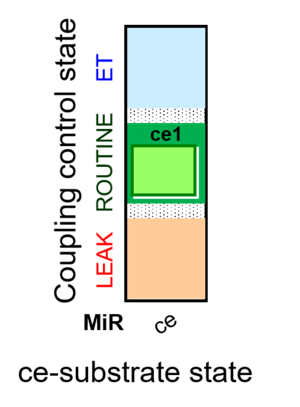 | A |
| SUIT-013 AmR ce D023 | O2 dependence of H2O2 production ce | 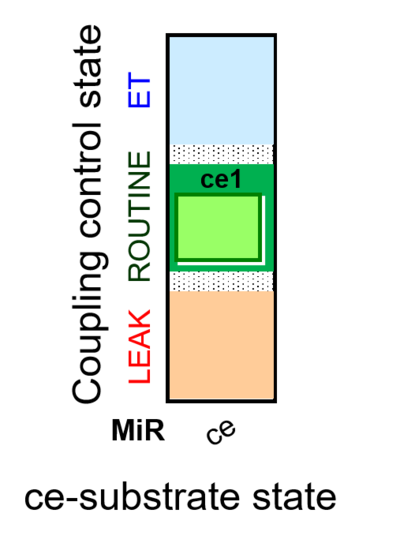 | A SUIT-013 |
| SUIT-014 | GM+P+S_OXPHOS+Rot_ET | 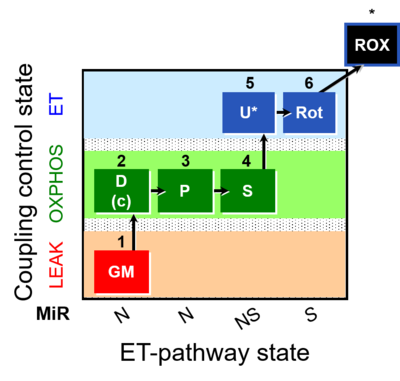 | A: Cells or tissue types that display a preference for GM over PM to support NADH-linked respiration |
| SUIT-014 O2 pfi D042 | NS(PGM) | 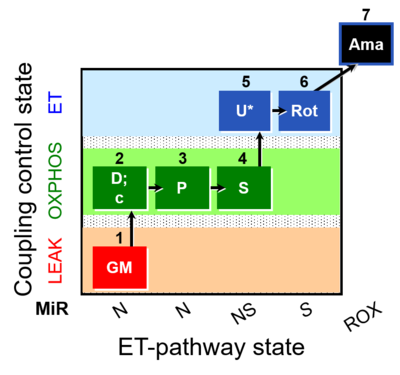 | A SUIT-014 |
| SUIT-015 | F+G+P+S_OXPHOS+Rot_ET | 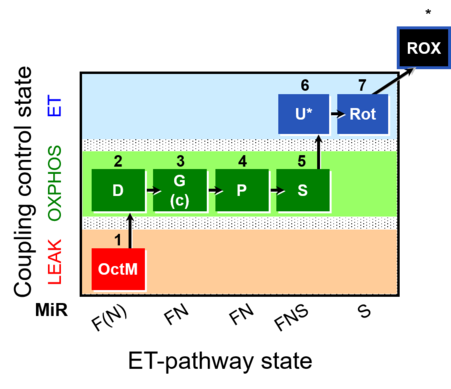 | A: F-pathway in LEAK state and OXPHOS state |
| SUIT-015 O2 pti D043 | FNS(Oct,PGM) | 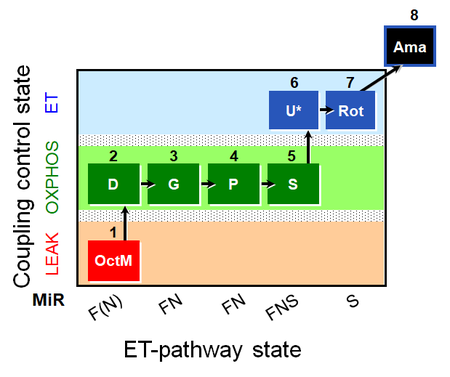 | A pti: permeabilized tissue- SUIT-015 |
| SUIT-016 | F+G+S+Rot_OXPHOS+Omy | 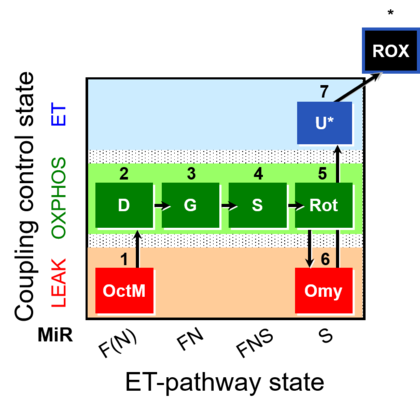 | A: F-pathway in LEAK state and OXPHOS state |
| SUIT-016 O2 pfi D044 | FNS(Oct,GM) | 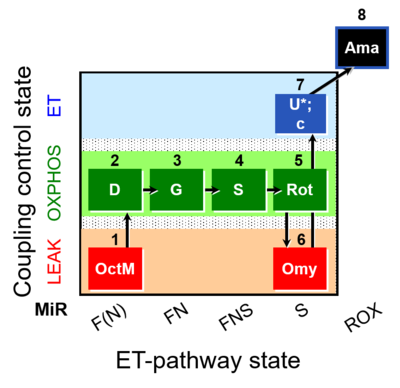 | A pfi: permeabilized fibers- SUIT-016 |
| SUIT-017 | F+G+S_OXPHOS+Rot_ET | 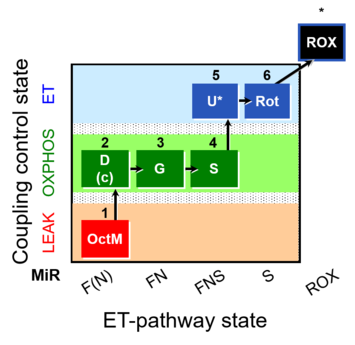 | A: |
| SUIT-017 O2 mt D046 | FNS(Oct,GM) | 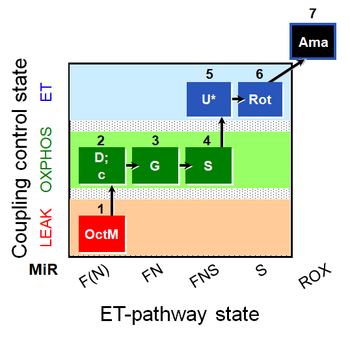 | A: SUIT-017 |
| SUIT-017 O2 pfi D049 | FNS(Oct,GM) |  | A: SUIT-017 |
| SUIT-018 | O2 dependence-H2O2 | 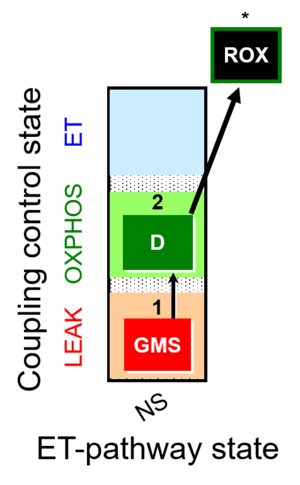 | A: A protocol for oxygen dependence of O2 flux and H2O2 flux in isolated mitochondria,tissue homogenate or permeabilized cells |
| SUIT-018 AmR ce-pce D068 | O2 dependence-H2O2 | 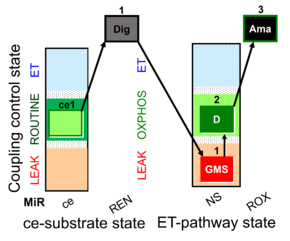 | A -SUIT-018 - SUIT protocol for simultaneous determination of O2 and H2O2 flux in permeabilized cells at various oxygen concentrations |
| SUIT-018 AmR mt D031 | O2 dependence-H2O2 | 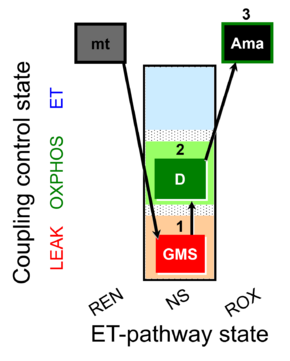 | A -SUIT-018 - SUIT protocol for simultaneous determination of O2 and H2O2 flux in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells) at low oxygen concentration (tissue normoxia) |
| SUIT-018 AmR mt D040 | NS(GM) |  | A: simultaneous determination of O2 and H2O2 flux in mitochondrial preparations (isolated mitochondria, tissue homogenate and permeabilized cells)-SUIT-018 |
| SUIT-018 AmR mt D041 | O2 dependence-H2O2 |  | A -SUIT-018 |
| SUIT-018 O2 mt D054 | 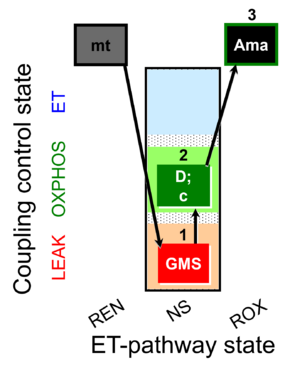 | B: SUIT-018 | |
| SUIT-019 | Pal+Oct+P+G_OXPHOS+S+Rot_ET | 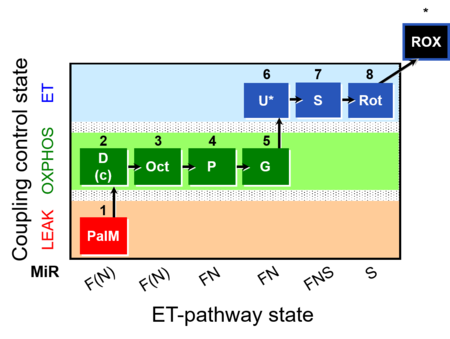 | A: |
| SUIT-019 O2 pfi D045 | FNS(PalOct,PGM) | 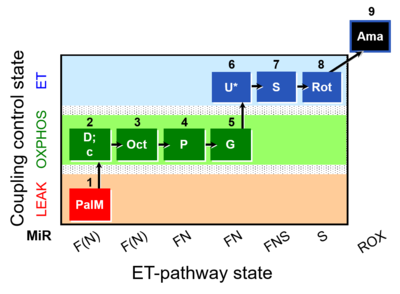 | A SUIT-019 |
| SUIT-020 | PM+G+S+Rot_OXPHOS+Omy | 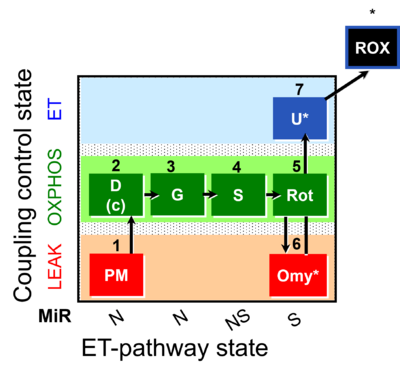 | A: simultaneous determination of O2 flux and mt-membrane potential |
| SUIT-020 Fluo mt D033 | NS(PGM) | 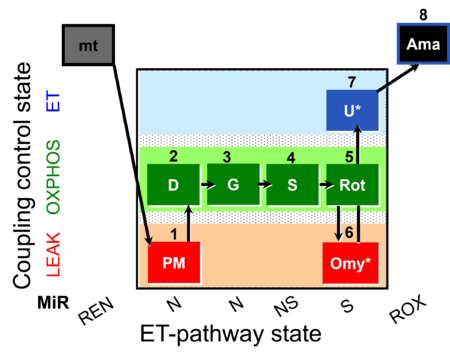 | A:short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)- SUIT-020 |
| SUIT-020 O2 mt D032 | Q-junction additivity and respiratory control for membrane potential | 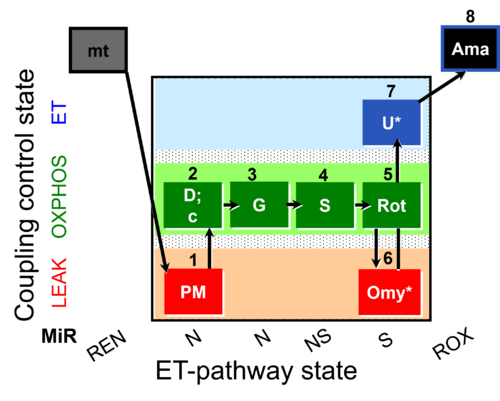 | A: short protocol for determination of O2 flux in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)-SUIT-020 |
| SUIT-021 | OXPHOS (GM+S+Rot+Omy) | 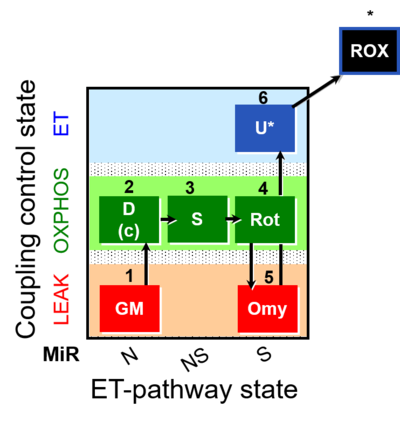 | A: simultaneous determination of O2 flux and mt-membrane potential |
| SUIT-021 Fluo mt D036 | NS(GM) | 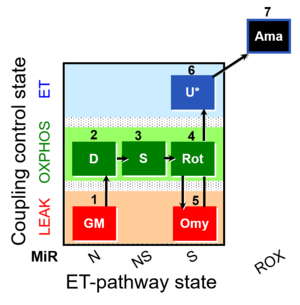 | A: short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permeabilized cells, and tissue homogenate)-SUIT-021 |
| SUIT-021 O2 mt D035 | NS(GM) | 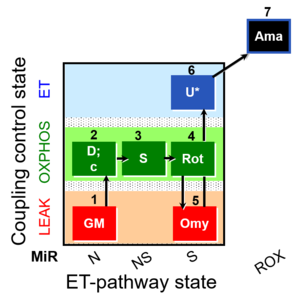 | A:short protocol for simultaneous determination of O2 flux and mitochondrial membrane potential in mitochondrial preparations (isolated mitochondria, permabilized cells, and tissue homogenate)-SUIT-021 |
| SUIT-022 | AOX (ce CN+SHAM) | 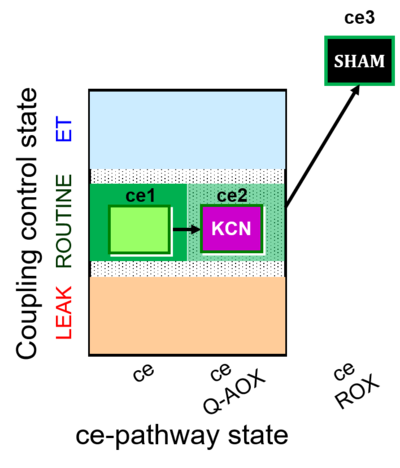 | A: Determination of the respiration due to the alternative oxidase pathway |
| SUIT-022 O2 ce D051 | AOX-ce CN+SHAM | 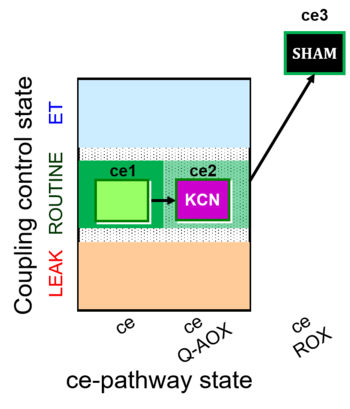 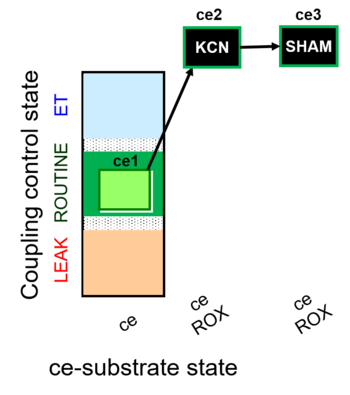 | A Determination of the respiration due to the alternative oxidase pathway -SUIT-022 |
| SUIT-023 | AOX-ce SHAM+CN | 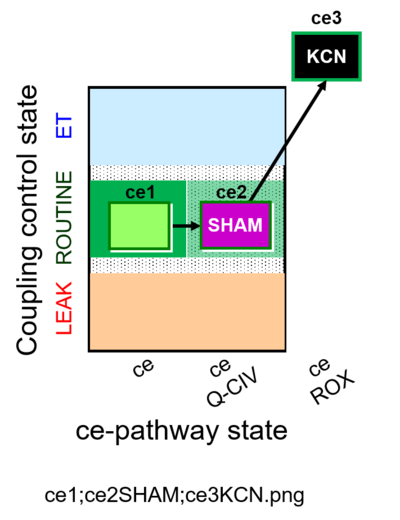 | A: Determination of respiration through the CIII-CIV pathway |
| SUIT-023 O2 ce D053 | AOX-ce SHAM+CN | 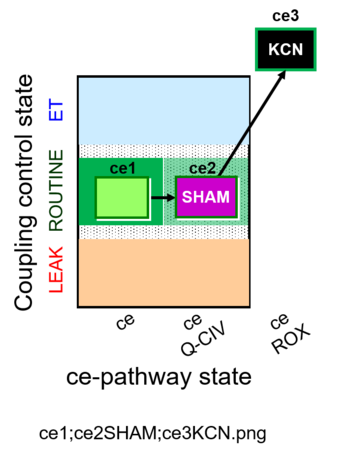 | A Determination of the respiration due to the cytochrome c oxidase pathway -SUIT-023 |
| SUIT-024 | ATPase (PM) | 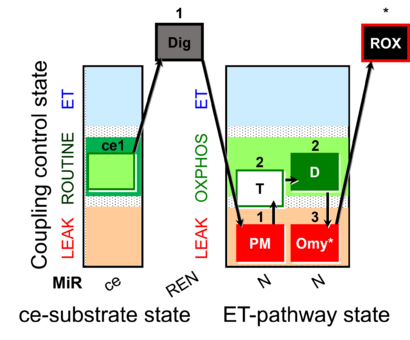 | A: Determination of ATPase activity in mitochondrial preparations. |
| SUIT-024 O2 ce-pce D056 | N(PM) | 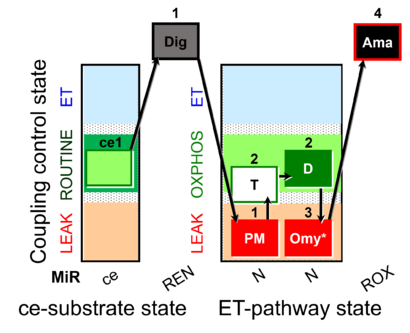 | Determination of the presence of ATPases in mitochondrial preparations - SUIT-024 |
| SUIT-025 | OXPHOS (F+M+P+G+S+Rot) | 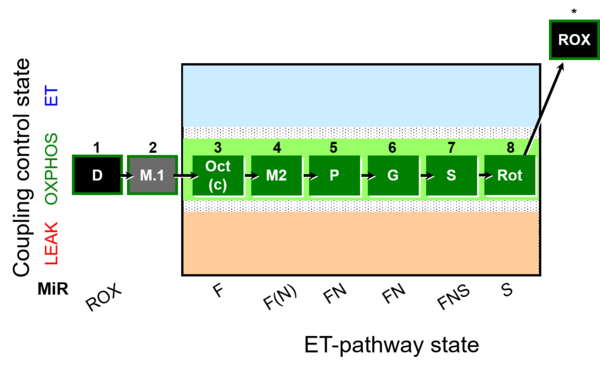 | |
| SUIT-025 O2 mt D057 | FNS(Oct,PGM) | 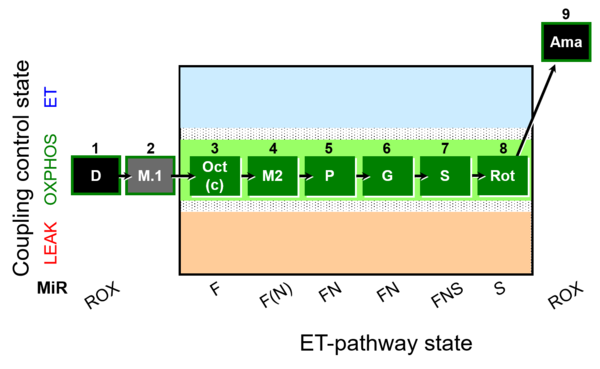 | - SUIT-025 |
| SUIT-026 | RET | 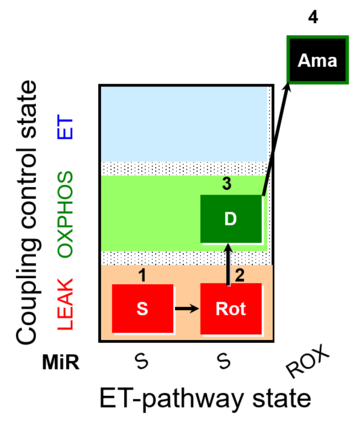 | A Short protocol to study RET-related H2O2 production |
| SUIT-026 AmR ce-pce D087 | RET | 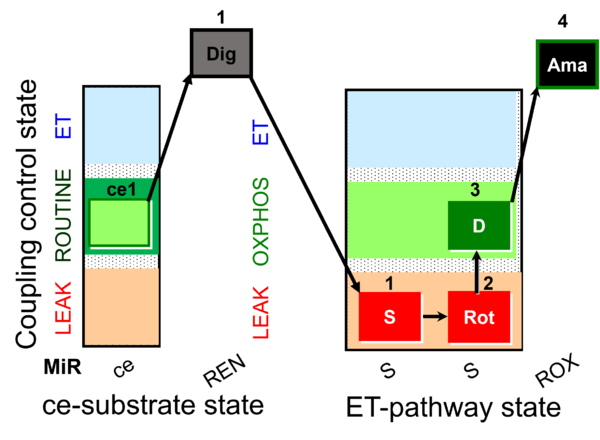 | A - SUIT-026 - Protocol for the evaluation of ROS production by RET in permeabilized cells |
| SUIT-026 AmR mt D064 | RET | 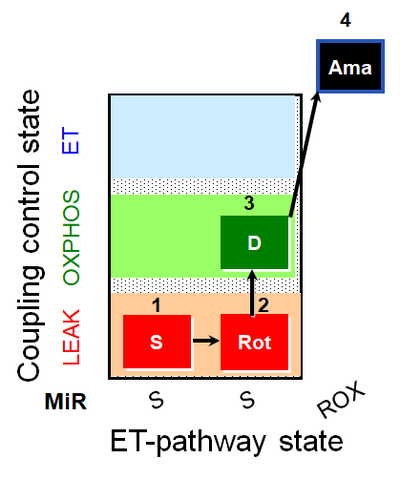 | A - SUIT-026 - Protocol for the evaluation of ROS production by RET in mtprep |
| SUIT-026 AmR mt D077 | RET |  | A - SUIT-026 - Protocol for the evaluation of ROS production by RET in mtprep |
| SUIT-026 O2 ce-pce D088 | RET (respiratory control) of SUIT-026 AmR ce-pce D087 | 400px | B - SUIT-026 - Protocol for the respiratory control of SUIT-026 AmR ce-pce D087 |
| SUIT-026 O2 mt D063 | RET (respiratory control) of SUIT-026 AmR mt D064 | 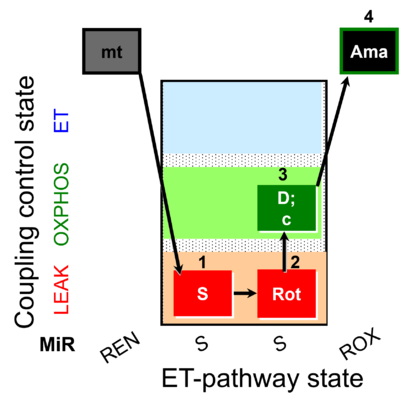 | B - SUIT-026 - Protocol for the respiratory control of SUIT-026 AmR mt D064 |
| SUIT-027 | Malate anaplerosis | 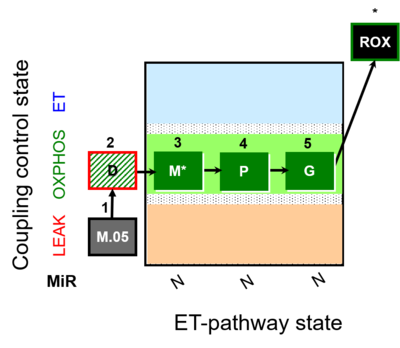 | A: Malate anaplerotic pathway |
| SUIT-027 O2 ce-pce D065 | Malate anaplerosis | 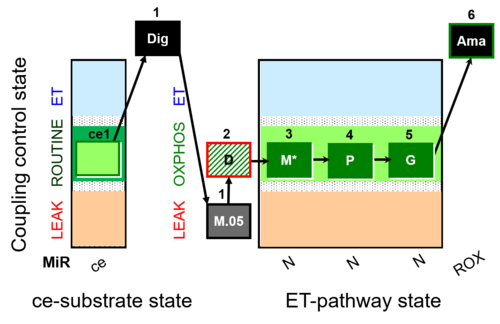 | A - SUIT-027 - Protocol for analysis of the malate anaplerotic pathway |
| SUIT-028 | NS(PGM) | 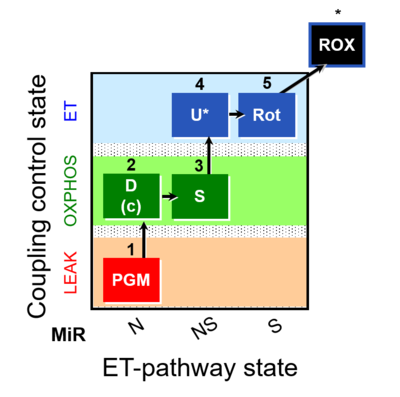 | C MiPNet18.13 IOC84 Alaska |
| SUIT-029 O2 mt D066 | QC_imt_PM_T+OXPHOS+c+Omy_ET_G+S+Rot | 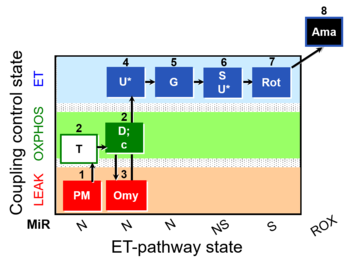 | A: quality control evaluation of mitochondrial preparations - SUIT-029 |
| SUIT-031 | PM+S+Rot | 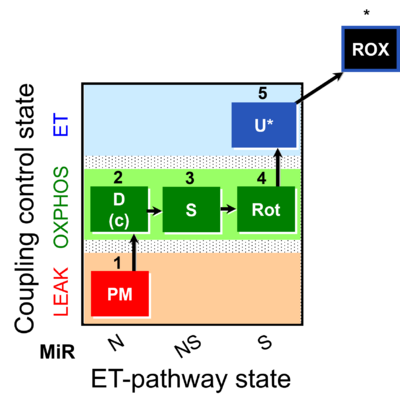 | A: |
| SUIT-031 O2 ce-pce D079 | PM+S+Rot | 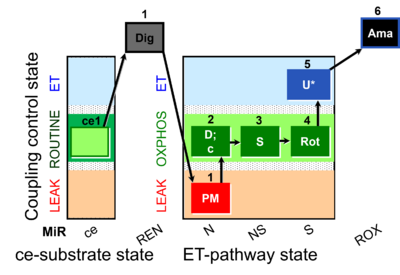 | A to analyse O2 flux in the NS- pathway control state in permeabilized cells- SUIT-031 |
| SUIT-031 O2 mt D075 | PM+S+Rot | 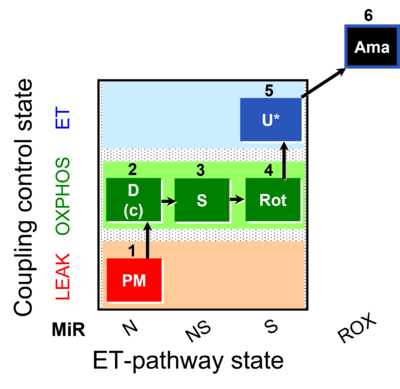 | A to analyse O2 flux in the NS- pathway control state on isolated mitochondria- SUIT-031 |
| SUIT-031 Q ce-pce D074 | PM+S+Rot | 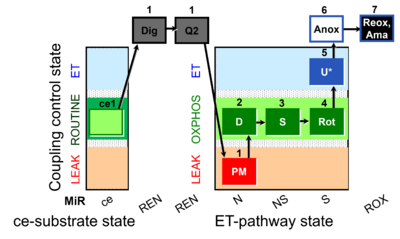 | A to analyse O2 flux and the Q-redox state in the NS- pathway control state on permeabilized cells- SUIT-031 |
| SUIT-031 Q mt D072 | PM+S+Rot | 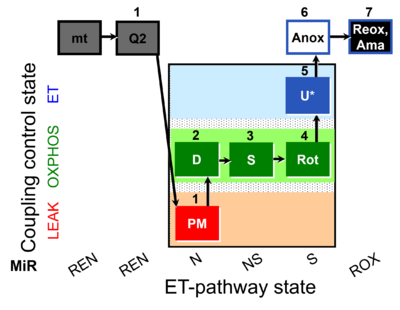 | A to analyse O2 flux and the Q-redox state in the N-,NS- and S- pathway control state on isolated mitochondria- SUIT-031 |
| SUITbrowser | Use the SUITbrowser to find the substrate-uncoupler-inhibitor-titration (SUIT) protocol most suitable for addressing your research questions.
Open the SUITbrowser: http://suitbrowser.oroboros.at/
 How to find a DL-Protocol (DLP) How to find a DL-Protocol (DLP) |


